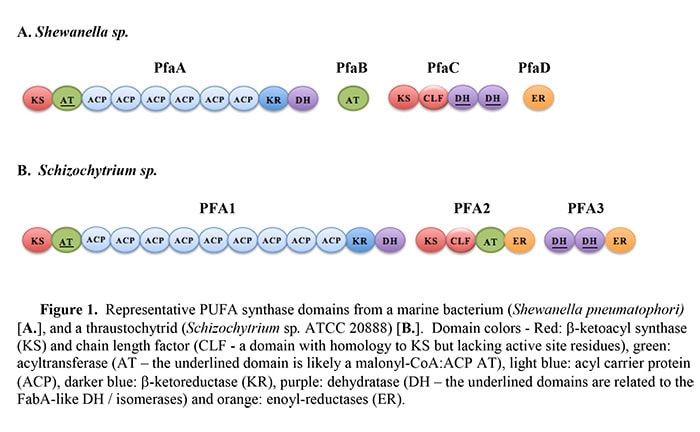
Polyunsaturated fatty acid (PUFA) synthases are Type I iterative enzymes that carry out de novo synthesis of specific long-chain PUFAs using malonyl-CoA as the carbon source. The products of these synthases include three PUFAs that are of primary interest to human health and nutrition: docosahexaenoic acid (DHA, 22:6 n-3), eicosapentaenoic acid (EPA, 20:5 n-3) and arachidonic acid (ARA, 20:4 n-6). PUFA synthases are primarily found in marine bacteria and the thraustochytrids, a group of marine heterokont microalgae. Recently, the range of microorganisms containing PUFA synthases has been expanded to include examples from fresh water bacteria [1], terrestrial myxobacteria [2, 3] and possibly another type of eukaryotic microalga (Emiliania huxleyi) [4]. The subunit and domain organization of examples of PUFA synthases from a marine bacterium and a thraustochytrid are shown in Figure 1. Unlike the “standard pathway” in which a series of elongase and desaturase reactions generate PUFAs from pre-existing, short-chain fatty acids, PUFA synthases do not have a requirement for molecular oxygen for insertion of the carbon-carbon double bonds and the synthesis mechanism is sometimes referred to as the ‘anaerobic pathway’.
PUFA synthases are often referred to as a polyketide synthase (PKS) system since several of their enzymatic domains have significant homology to those typically found in Type I PKSs. But ‘PKS-like’ would be more appropriate since PUFA synthases possess unique features that distinguish them from previously described systems. They are hybrid systems incorporating elements of Type I PKS and of Type II PKS and FAS systems. For example, several of the enzymatic domains embedded in the multi-domain subunits of PUFA synthases are homologous to those typically found only as discreet components of Type II PKS systems or of Type II fatty acid synthases (FASs). These include two tandem domains with homology to a dehydratase/isomerase (DH/I) found in some Type II FAS systems, the FabA-like enzymes, and a tandem pair of domains with homology to the β-ketoacyl synthase (KS) – chain-length factor (CLF) heterodimeric enzyme found in Type II PKS systems. CLFs are proteins with homology to KSs but lacking the active site residues. Additionally, all PUFA synthases identified to date contain from 3 to 10 tandem acyl-carrier protein (ACP) domains. Thorough reviews of various aspects of PUFA synthase biochemistry, their homologs and potential phylogenetic relationships can be found in [5] and in [6].
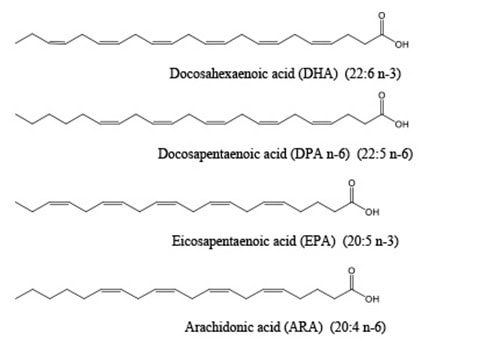
PUFA synthases produce a limited set of fatty acids as their primary products, including DHA, EPA and docosapentaenoic acid (DPA n-6, 22:5 n-6). This list was recently expanded to include arachidonic acid (ARA, 20:4 n-6) being produced in a marine bacterium, Aureispira marina [7] as well as linoleic acid in a myxobacterium (LA, 18:2 n-6) [2]. Most of these fatty acids are typically associated with primary metabolism in animals rather than the secondary metabolites normally produced by PKS systems. Depending on the specific PUFA synthase, the main product can be a single PUFA such as DHA, EPA, or ARA, or a mix of PUFAs such as DHA and DPA n-6 or DHA and EPA. All of these fatty acids are 18 to 22 carbons in length and all have between 2 and 6, methylene interrupted, carbon-carbon double bonds (some examples are shown in Figure 2). The double bonds are always in the cis configuration and the first double bond occurs at either the n-3 or n-6 position. The important roles of some of these PUFAs, especially DHA, EPA and ARA, in human physiology have been well documented. Why these particular PUFAs should be made and accumulate in microorganisms remains an open question. Evidence that PUFAs play essential roles in some of these microorganisms was demonstrated by the dependence on supplemental PUFAs for growth of strains of Schizochytrium sp. ATCC 20888 in which the PUFA synthase had been inactivated [8]. Additionally, the potential roles for PUFAs in marine bacteria, including anti-oxidative functions and involvement of PUFA synthases in production of a very long chain polyunsaturated hydrocarbon (C31:9) are discussed in detail in [6].
Figure 2. Structures of some of the fatty acids produced by PUFA synthases
Bacterial PUFA synthases
Genes encoding PUFA synthases were first discovered in marine bacteria. Contrary to prevailing views of the time, DeLong et al. [9] established that certain marine bacteria contained significant levels of PUFAs, such as EPA and DHA. In a remarkable achievement, Yazawa [10] was able to use a cosmid derived from one of those strains (Shewanella strain SCRC-2378, presently identified as Shewanella pneumatophori) to produce EPA in E. coli. EPA was the only new fatty acid produced in the E. coli transformants and subsequent work revealed that five open reading frames (ORFs) present in the cosmid were necessary and sufficient for EPA accumulation in the heterologous host [11]. It was then proposed in 2001 that the proteins encoded by four of these ORFs represented subunits of a novel Type I enzyme capable of de novo PUFA synthesis [12]. This hypothesis was based on a number of factors, including: examination of the domain functions, in vitro activity assays and the observation that PUFA synthesis could occur in the absence of O2. The fifth ORF, identified as essential for EPA synthesis, was found to encode an accessory enzyme required for activation of the ACP groups of the PUFA synthase enzyme by attachment of a co-factor by a phosphopantetheinyl transferase (PPTase). Although E. coli contains several endogenous PPTases, they were apparently unable to recognize and activate the ACP domains of the Shewanella PUFA synthase.
Homologous gene sets encoding PUFA synthase subunits have been identified in numerous other marine bacteria [1, 2, 13, and the comprehensive reviews in 5 and 6]. In several cases, these homologs have been confirmed as being PUFA synthases by heterologous expression and detection of novel PUFAs in the new host, including the production of ARA and LA. While variation in the domain content and organization has been observed, including additional putative KS and FabA-like DH domains, the key distinguishing features are still apparent in the verified PUFA synthases. These examples indicate that most of PUFAs produced in bacteria are likely the products of PUFA synthase systems.
Eukaryotic PUFA synthases
Concurrent with the proposal of a bacterial PUFA synthase system, a homologous system was characterized in a eukaryotic microalga that had been developed as a commercial source of oil enriched in DHA: Schizochytrium sp. ATCC 20888 [12, 14]. Schizochytrium sp. is a member of the thraustochytrids, a group of large-celled heterokont protists [15]. Subsequently, PUFA synthase genes have been found in several other Thraustochytrids (e.g., see [16] and [17]). Presumably the thraustochytrids acquired PUFA synthase genes from marine bacteria via lateral gene transfer. The domains of the thraustochytrid PUFA synthases are organized somewhat differently than most of the bacterial versions (Figure 1). Typically there are three subunits, instead of four, and there is a duplication of the enoyl-reductase (ER) domain. While the bacterial PUFA synthase subunit genes are organized as part of an operon, separate genes encode the subunits of the eukaryotic thraustochytrid synthases. It is noteworthy that, in the cases where it has been examined, PFA1 and 2 are adjacent in a head-to-head arrangement. Several thraustochytrids that posses PUFA synthases have also been shown to contain a partial, or complete, set of enzymes of the standard PUFA synthesis pathway (i.e., the elongase and desaturase enzymes). In some cases the standard pathway is functional, including production of DHA [16]. In the case of Schizochytrium sp. ATCC 20888, a partial set of standard pathway enzymes was identified but a critical Δ12 desaturase activity is missing and this system is therefore not capable of synthesizing PUFAs from the shorter saturated fatty acid products of the endogenous FAS [8]. Furthermore, inactivation of the PUFA synthase in this organism results in auxotrophy and supplementation of the medium with PUFAs is required for growth [8].
Genes that encode proteins with homology to PUFA synthases continue to be identified. A recent example was the discovery of a homolog in the eukaryotic coccolithophore, Emiliania huxleyi [4]. In this case, all of the domains of the putative PUFA synthase were encoded on a single large protein. E. huxleyi is a major component of the of the global phytoplankton community and is known to produce significant levels of PUFA. As in the case of other eukaryotes that possess a PUFA synthase system, E. huxleyi also contains a suite of elongase and desaturase enzymes of the standard PUFA synthesis pathway [18]. It has yet to be determined what the relative contributions of the two pathways for synthesis of the PUFAs in this organism may be.
Potential PUFA synthesis mechanisms
PUFA synthases are Type I iterative systems and some of the synthesis steps can be inferred by comparison to similar systems. The products of PUFA synthases are linear fatty acids that are 18, 20 or 22 carbons in length. These are built-up by sequential addition of two carbons at a time, derived from malonate, requiring 8, 9 or 10 condensation reactions, respectively. The PUFA products contain between 2 and 6 carbon-carbon double bonds and therefore some of the extension cycles require full reduction of the keto group formed on the β-carbon by the condensation reaction. All PUFA synthases contain a set of domains typically associated with this type of extension and full reduction cycle: malonyl-CoA:ACP transacylase (MAT), ACP, KS, β-ketoreductase (KR), DH and ER (Figure 1). Most of these domains are easily recognized via homology searches, but identification of the DH domain has been less clear. This putative domain occurs just downstream of the KR domain of PFA1 (PfaA in bacterial PUFA synthases). An analysis of the last ~300 amino acids of that subunit using the Pfam program [19] identifies this region as containing a domain of the PKS-DH family. In addition, a motif that has been associated with some PKS-DH domains (L/IxxHxxxGxxxxP) [20] is typically present in this region. A domain with these characteristics is found in a similar region of all known PUFA synthases. Although it has yet to be demonstrated, this domain could account for the dehydration reactions in those elongation cycles that are associated with a full reduction of the newly added β-carbon.
While domains indicated above can account for a full reduction-extension cycle, other aspects associated with PUFA synthesis are much less clear. In particular, as discussed below, there are unresolved questions associated with the type and placement of the carbon-carbon double bonds, the key factors that influence the end product profiles, the role(s) of the multiple tandem ACP domains and the off-loading of the end products from the synthase.
Carbon-carbon double bonds
While chain extension cycles add two carbons at a time, the double bonds in the products of PUFA synthases occur in three carbon intervals. Additionally, while the DH reactions associated with PKS and FAS systems insert carbon-carbon double bonds in the trans configuration, the carbon-carbon double bonds in the products of PUFA synthases are always in the cis configuration. A scheme for a sequence of reactions that could result in production of a PUFA (EPA) has been described ([12 – Supplemental information] and redrawn in [21]). In this scheme, the double bonds are inserted as part of the normal chain extension cycle and at the appropriate times are either reduced to yield the saturated β-carbon, or undergo trans to cis isomerizations. For correct localization of the double bonds, the isomerizations would need to occur with 2,3 bond migration in some cases and without bond migration in other cases (an in situ – 2,2 – isomerization). Although examples of enzymes capable of in situ isomerizations have been described, they are rare and domains present in PUFA synthases do not show homology to those particular enzymes. Other reaction schemes resulting in methylene interrupted cis double bonds can be envisioned and this remains a major unresolved question.
Although the precise mechanism for double bond creation and positioning is unclear, it is highly likely that it will involve the tandem domains that have homology to FabA-like enzymes. FabA enzymes are associated with some Type II fatty acid synthesis systems of bacteria and in addition to a DH activity that introduces a double bond in the linear carbon chain, those same enzymes can also carry out trans-cis isomerization, with 2,3 migration, of that double bond [22]. In E. coli, FabA is needed for formation of unsaturated fatty acids (e.g., cis-vaccenic acid, 18:1 n-7) and inactivation of the FabA gene results in auxotrophy that requires supplementation with unsaturated fatty acids for growth.
End product profiles
Several thraustochytrid PUFA synthases produce DPA n-6 in addition to DHA (e.g., see [12] and [17]). Subunit and domain substitution experiments between homologous PUFA synthases from two thraustochytrid species were used to demonstrate that the ratio of the n-3 to n-6 fatty acid products could be influenced by activities associated with a FabA-like domain of PFA3. Specifically, when the second FabA-like domain of the PUFA synthase from Schizochytrium sp. ATCC 20888 was replaced with a homologous region from a system that produced primarily DHA (derived from Thraustochytrium 23B), the ratio of DHA to DPA n-6 produced in the hybrid system was altered from ~2.4:1 of the native synthase to ~10:1 in the hybrid synthase [17]. While the precise mechanism is unclear, this again points to the involvement of at least one of the FabA-like domains in the formation and placement of the cis double bonds.
There are several examples of bacterial PUFA synthases that produce essentially a single product, such as EPA or DHA. Orikasa et al. [23] examined the synthases from Shewanella pneumatophori and from Moritella marina MP-1 that produce EPA and DHA, respectively. Based on co-expression and subunit substitution experiments performed in E. coli, they provided evidence that the PfaB subunit of these systems can influence EPA vs. DHA end product formation. The PfaB subunit of bacterial PUFA synthases contain an AT domain and a homolog of this domain is also embedded in a multi-domain subunit in thraustochytrid PUFA synthases (Figure 1). No other specific role has been ascribed to this AT domain at this time.
Of particular interest for end product chain length considerations are the tandem KS-CLF domains that are present in all PUFA synthases. The CLF domain has sequence homology to KS domains but lacks the active site cysteine required for the condensation reaction. The KS – CLF heterodimer is a key feature of aromatic Type II PKS systems and the CLF domain has been suggested to play a key role in determination of the chain length of the final products [24]. The tandem KS – CLF domains are a distinguishing feature of the PUFA synthase systems and their resemblance to the Type II PKS domains suggests that they may play a role in chain length determination. As is the case for several aspects of the PUFA synthesis reaction scheme, this hypothesis awaits experimental examination.
Tandem ACP domains
Another distinguishing feature of PUFA synthases is the presence of multiple tandem ACP domains with the current range being 3 to 10. Two or three tandem ACP domains have been observed in some Type I modular PKS systems. For example, in the curacin synthesis system, a mixed PKS/non-ribosomal peptide synthetase system, one of the modules contains three tandem ACP domains [25]. Several enzymatic activities in addition to those associated with elongation cycle reactions occur in that module. The presence of multiple ACP domains appears to enhance the efficiency of the reactions associated with that module. Each of the individual ACP domains could support all of the reactions of the module, but the overall efficiency of the reactions was enhanced by the presence of the other, active domains [25]. The tandem ACP domains may play a similar role in PUFA synthases. Experiments using the Shewanella japonica PUFA synthase revealed that each of that system’s six ACPs could function by itself to support production of the normal product – EPA [26]. Also, the presence of a greater number of functional ACP domains resulted in a higher rate of accumulation of the EPA product [26]. Any function of the multiple ACPs, beyond increasing the productivity of the system, is not apparent. Recent work has indicated that the ACP domains of the Photobacterium profundum PUFA synthase, and presumably in other PUFA synthases, are arranged like ‘beads on a string’, again suggesting that the ACP domains operate in an independent fashion [27].
The sequences of the ACP domains of PUFA synthases are highly conserved, especially in the central part of the domain surrounding the serine residue to which pantetheine is covalently linked. When expressing a PUFA synthase in a heterologous host it may be necessary to also provide a PPTase capable of recognizing those ACP domains. Several PPTases have been identified that are capable of this task: e.g., HetI, from the related hgl (heterocyst glycolipid) synthesis system of cyanobacteria [28], as well as Sfp from Bacillus and Svp from Streptomyces verticillus [26].
Chain termination / off-loading mechanisms
As for many Type I iterative systems, the method of chain termination for most PUFA synthases is not evident in the primary structure. Expression of PUFA synthases, and an appropriate PPTase, in heterologous hosts without co-expressing additional genes has resulted in PUFA accumulation in those hosts. These results indicate the off-loading mechanisms are either inherent to the system or that factors already present in the host can facilitate the off-loading. Evidence suggests that different off-loading mechanisms may be in place for PUFA synthase systems from different types of organisms. When PUFA synthases from marine bacteria are heterologously expressed in other bacteria the products of the synthase are found in the phospholipid fraction of the new host [11]. As previously mentioned, PUFA synthases contain a second AT domain that has been implicated in end product determination in some bacterial systems [23]. It is possible, that this AT domain could be associated with off-loading of the end products from the ACP domains of the PUFA synthases and subsequent incorporation into the phospholipids of the heterologous host. Interestingly, a domain with homology to glycerolipid acyltransferases, most likely a 1-acylglycerol-3-phosphate O-acyltransferase (AGPAT), is a part of PUFA synthases isolated from myxobacteria [2]. The presence of this type of domain, integrated into one of the multi-domain subunits, provides strong evidence for a direct transfer of the fatty acid products to glycerol lipids in these systems [2, 3]. In contrast to the above bacterial cases, when the Schizochytrium sp. ATCC 20888 PUFA synthase was expressed in E. coli, DHA and DPA n-6 accumulated as free fatty acids [28]. Free fatty acids were also detected as the products of in vitro assays of extracts of E. coli expressing the Schizochytrium PUFA synthase [28]. This implies involvement of thioesterase activities, either as a part of an intrinsic domain or by factors outside of the PUFA synthase structure. Recently, two proteins identified in extracts of Schizochytrium sp. ATCC 20888 were shown to be able to enhance in vitro activity of the Schizochytrium PUFA synthase [29]. Both of these proteins had homology to a specific type of thioesterase (the 4-hydroxybenzoyl-CoA-like thioesterases) and showed maximal enhancing activity when expressed together. Inactivation of the genes in Schizochytrium encoding these proteins resulted in a decrease, but not a total loss, of PUFA production in those mutant lines, suggesting that more than one off-loading mechanism could be functioning in this system.
Ecological roles and commercial applications
The benefits of long chain PUFAs in human health have been well established and it is recognized that a key source for these fatty acids are microorganisms from the marine environment. These microorganisms are a critical source for most of the PUFAs found in the food chain and are passed to the fish and other marine animals that are harvested by humans. The contribution of the PUFA synthase pathway, versus the standard PUFA synthesis pathway, in the production of these fatty acids has not been determined but could be significant. It now appears that most of the PUFAs found in marine bacteria are products of the PUFA synthesis pathway. Many marine protists of the thraustochytrid group have also been shown to contain the PUFA synthase genes. In several cases the PUFAs present in those algae have been confirmed to be the products of the PUFA synthases and new organisms harboring the system continue to be discovered. A recent example is the detection of a PUFA synthase homolog in Emiliania huxleyi, which is a major component of the marine food chain, and its oils can contain high levels of PUFAs. E. huxleyi also contains a full complement of the standard PUFA synthesis pathway enzymes [18] and it has yet to be determined if its PUFAs are the products of one or the other system, or of both.
The first eukaryotic organism in which the PUFA synthase was discovered was Schizochytrium sp. ATCC 20888 [12]. This organism had been isolated based on growth and PUFA productivity characteristics [14]. It was only after commercialization of the organism’s biomass and extracted oil that it was discovered that its PUFAs, DHA and DPA n-6, were the exclusive products of a PUFA synthase system. Other thraustochytrid strains that utilize the PUFA synthase system have been developed as commercial sources of PUFA enriched oils. These organisms are capable of reaching very high cell densities with high oil and PUFA contents [14]. The lack of a requirement for oxygen to form the many double bonds in the end-product PUFAs as well as the overall lower energy requirement for PUFA synthesis may provide an advantage when producing these oils via fermentation.
Although much remains to be resolved concerning mechanistic aspects of the PUFA synthase synthesis reactions and the accumulation of the products in host organisms, some progress with potential commercial relevance has been made. For example, specific domains that influence EPA versus DHA formation in bacterial synthases have been identified [23]. In the thraustochytrid synthase, replacement of a single domain significantly increased the production of DHA with a corresponding reduction in DPA n-6 levels while retaining the overall growth characteristics of the parent strain [17]. It is likely that additional opportunities for directed modification of the PUFA synthase and accessory enzyme will emerge as the reaction mechanisms become more clearly defined.
Another area of interest is the production of PUFAs in oilseed crop plants as an alternative to harvesting of marine fishes or growth of algae via sugar-fed fermentation. PUFAs production in oilseed crop plants could have significant cost and sustainability benefits versus the current methods of production. Commercially developed oilseed crops typically do not contain PUFAs with chain lengths longer than 18 carbons and with more than three double bonds. Considerable progress has been made in recent years using genes encoding enzymes of the standard PUFA synthesis pathway to modify the endogenous plant fatty acids to yield PUFA end-products including some oils with EPA and DHA levels similar to those found in fish oils [30]. The use of genes encoding a PUFA synthase system to produce these fatty acids in Canola has also been demonstrated [31]. While the levels of DHA reported using the PUFA synthase pathway are currently lower than those achieved using the standard pathway, the de novo synthesis aspect allows for the potential production of PUFAs in a variety of plant backgrounds.
Due to the relatively recent discovery of PUFA synthases, there are still many questions to answer in these unique and important biosynthesis systems. One of the key, open questions revolves around the relative contribution of these systems to PUFAs found in the environment. A second key area is the biosynthetic mechanisms that control end-product formation. The continuing and increasing interest in PUFAs’ roles in human health along with the need for novel and sustainable production will continue to drive new discoveries in the years to come.
References
- Dailey, F.E., McGraw, J.E., Jensen, B.J., Bishop, S.S., Lokken, J.P., Dorff, K.J., Ripley, M.P. and Munro, J.B. The microbiota of freshwater fish and freshwater niches contain omega-3 fatty acid-producing Shewanella species. Appl. Environ. Microbiol. 82:218–231 (2015) (DOI: 10.1128/AEM.02266-15).
- Gemperlein, K., Rachid, S., Garcia, R.O., Wenzel, S.C. and Muller, R. Polyunsaturated fatty acid biosynthesis in myxobacteria: Different PUFA synthases and their product diversity. Chem Sci. 5:1733–1734 (2014) (DOI: 10.1039/c3sc53163e).
- Gemperlein, K., Zipf, G., Bernauer, H.S., Muller, R. and Wenzel, S.C. Metabolic engineering of Pseudomonas putida for production of docosahexaenoic acid based on a myxobacterial PUFA synthase. Metab. Eng. (2015) (DOI: 10.1016/j.ymben.2015.11.001).
- Sasso, S., Pohnert, G., Lohr, M., Maria Mittag, M. and Hertweck, C. Microalgae in the postgenomic era: A blooming reservoir for new natural products (section on polyunsaturated fatty acids). FEMS Microbiol. Rev. 36, 761–785 (2012) (DOI: 10.1111/j.1574-6976.2011.00304).
- Shulse, C.N. and Allen, E.E. Widespread occurrence of secondary lipid biosynthesis potential in microbial lineages. PLoS ONE 6(5) (2011) e20146 (DOI: 10.1371/ journal.pone.0020146).
- Yoshida, K., Hashimoto, M., Ryuji Hori, R., Adachi, T., Okuyama, H., Orikasa, Y., Nagamine, T., Shimizu, S., Ueno, A. and Morita, N. Bacterial Long-Chain Polyunsaturated Fatty Acids: Their Biosynthetic Genes, Functions, and Practical Use. Mar. Drugs, 14, 94 (2016) (DOI: 10.3390/md14050094).
- Ujihara, T., Nagano, M., Wada, H. and Mitsuhashi, S. Identification of a novel type of polyunsaturated fatty acid synthase involved in arachidonic acid biosynthesis. FEBS Letters 588, 4032–4036 (2014) (DOI: 10.1016/j.febslet.2014.09.023).
- Lippmeier, J.C., Crawford, K.S., Owen, C.B., Rivas, A.A., Metz, J.G. and Apt, K.E. Characterization of both polyunsaturated fatty acid biosynthetic pathways in Schizochytrium sp. Lipids 44:621–630 (2009) (DOI: 10.1007/s11745-009-3311-9).
- Delong, E.F. and Yayanos, A.A. Biochemical function and ecological significance of novel bacterial lipids in deep-sea procaryotes. Appl. Environ. Microbiol. 51(4), 730–737 (1986).
- Yazawa, K. Production of eicosapentaenoic acid from marine bacteria. Lipids 31, S-297 (1996) (DOI: 10.1007/BF02637095).
- Yu, R., Yamada, A., Watanabe, K., Yazawa, K., Takeyama, H., Matsunaga, T., and Kurane, R. Production of eicosapentaenoic acid by recombinant marine cyanobacterium, Synechococcus sp. Lipids 35, 1061–1064 (2000) (DOI: 10.1007/s11745-000-0619-6).
- Metz, J.G., Roessler, P., Facciotti, D, Levering, C., Dittrich, F., Lassner, M., Valentine, R., Lardizabal, K., Domergue, F., Yamada, A., Yazawa, K., Knauf, V. and Browse, J. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science, 293, 290–293 (2001) (DOI: 10.1126/science.1059593).
- Tanaka, M., Ueno, A., Kawasaki, K., Yumoto, I., Ohgiya, S., Hoshino, T., Ishizaki, K., Okuyama, H. and Morita, N. Isolation of clustered genes that are notably homologous to the eicosapentaenoic acid biosynthesis gene cluster from the docosahexaenoic acid-producing bacterium Vibrio marinus strain MP-1. Biotechnol. Lett. 21:939–945 (1999) (DOI: 10.1023/A:1005601606929).
- Barclay, W., Weaver, C., Metz, J. and Hansen, J. Development of a docosahexaenoic acid production technology using Schizochytrium: Historical perspective and update. Single cell oils: microbial and algal oils, Zvi Cohen and Colin Ratledge, Eds., 2nd ed. Chapter 4 (2010).
- Cavalier-Smith, T., Allsopp, M.T.E.P., Chao, E.E. Thraustochytrids are chromists, not fungi: 18 s rRNA signatures of heterokonta. Philos. Trans. R. Soc. Lond B, 346:387–397 (1994) (DOI: 10.1098/rstb.1994.0156).
- Matsuda. T., Sakaguchi, K., Hamaguchi, R., Kobayashi, T., Abe, E., Hama, Y., Hayashi, M., Honda, D., Okita, Y., Sugimoto, S. Okino, N. and Ito, M. Analysis of D-12-fatty acid desaturase function revealed that two distinct pathways are active for the synthesis of PUFAs in T. aureum ATCC 34304. J. Lipid Res. 53, 1210–1222 (2012) (DOI: 10.1194/jlr.M024935).
- Weaver, C.A., Zirkle, R., Doherty, D.H. and Metz, J.G. Chimeric PUFA polyketide synthase systems and uses thereof. US Patent 8003772 B2 (Issued 2011).
- Sayanova, O., Haslam, R.P., Calerón, M.V., López, N.R., Worthy, C., Rooks, P., Allen, M.J. and Napier, J.A. Identification and functional characterisation of genes encoding the omega-3 polyunsaturated fatty acid biosynthetic pathway from the coccolithophore Emiliania huxleyi. Phytochemistry, 72, 594–600 (2011).
- Finn, R.D., Coggill, P., Eberhardt, R.Y., Eddy, S.R, Mistry, J., Mitchell, A.L., Potter, S.C., Punta, M., Qureshi, M., Sangrador-Vegas, A., Salazar, G.A., Tate, J. and Bateman, A. The Pfam protein families database: Towards a more sustainable future. Nucl. Acids Res. 44 (D1) D279–D285 (2016) (DOI: 10.1093/nar/gkv1344).
- Donadio, S. and Katz, L. Organization of the enzymatic domains in the multifunctional polyketide synthase involved in erythromycin formation in Saccharopolyspora erythraea. Gene 111(1):51–60 (1992).
- Kaulmann, U. and Hertweck, C. Biosynthesis of polyunsaturated fatty acids by polyketide synthases. Angew Chem. Int. Ed. 41, 1866–1869 (2002) (DOI: 10.1002/1521-3773(20020603)41:11<1866::AID-ANIE1866>3.0.CO;2-3).
- Heath, R.J. and Rock, C.O. Roles of the FabA and FabZ b-hydroxyacyl-acyl carrier protein dehydratases in Escherichia coli fatty acid biosynthesis. J. Biol. Chem. 271, 27795–27801 (1996).
- Orikasa, Y., et al. pfaB products determine the molecular species produced in bacterial polyunsaturated fatty acid biosynthesis. FEMS Microbiol. Lett. 295, 170–176 (2009).
- Tang, Y., Tsai, S-C., and Khosla, C. Polyketide chain length control by chain length factor. J. Am. Chem. Soc. 125, 12708–12709 (2003) (DOI: 10.1021/ja0378759).
- Gu, L., Eisman, E.B., Dutta, S., Franzmann, T.M., Walter S., et al. Tandem acyl carrier proteins in the curacin biosynthetic pathway promote consecutive multienzyme reactions with a synergistic effect. Angewandte Chemie International Edition, 50, 2795–2798 (2011) (DOI: 10.1002/anie.201005280).
- Jiang, H., Zirkle, R., Metz, J.G., Braun, L., Richter, L., Van Lanen, S.G. and Shen, B. The role of tandem acyl carrier protein domains in polyunsaturated fatty acid biosynthesis. Am. Chem. Soc. 130, 6336–6337 (2008) (DOI: 10.1021/ja801911t).
- Trujillo, U., Vázquez-Rosam E., Oyola-Robles, D., Stagg, L.J., Vassallo, D.A., Vega, I.E., Arold, S.T., Baerga-Ortiz, A. Solution structure of the tandem acyl carrier protein domains from a polyunsaturated fatty acid synthase reveals beads-on-a-string configuration. PLoS One. 8(2):e57859 (2013) (DOI: 10.1371/journal.pone.0057859).
- Metz, J.G., Kuner, J., Rosenzweig, B., Lippmeier, J.C., Roessler, P., and Zirkle, R. Biochemical characterization of polyunsaturated fatty acid synthesis in Schizochytrium: Release of the products as free fatty acids. Plant Physiol. Biochem. 47, 472–478 (2009) (DOI: 10.1016/j.plaphy.2009.02.002).
- Metz, J.G., Kuner, J.M., McCaskill, D.G., and Foster, M.L. Factors for the production and accumulation of polyunsaturated fatty acids (PUFAs) derived from PUFA synthases. PCT/US2015/013274 WO 2015/116671 A3 (publication date; Sep 24, 2015).
- Ruiz-Lopez, N., Usher, S., Sayanova, O.V., Napier, J.A., and Haslam, R.P. Modifying the lipid content and composition of plant seeds: Engineering the production of LC-PUFA. Appl. Microbiol. Biotechnol. 99, 143–154 (2015) (DOI: 10.1007/s00253-014-6217-2).
- Walsh, T.A., Bevan, S.A., Gachotte, D.J., Larsen, C.M., Moskal, W.A, Owens Merlo, P.A., Sidorenko, L.V., Hampton, R.E., Stoltz, V., Pareddy, D., Anthony, G.I., Bhaskar, P.B., Marri, P.R., Clark, L.M., Chen, W., Adu-Peasah, P.S., Wensing, S.T., Zirkle, R. and Metz, J.G. Canola engineered with a microalgal polyketide synthase-like system produces oil enriched in docosahexaenoic acid. Nature Biotechnol. 34, 881–887 (2016) (DOI: 10.1038/nbt.3585)
Related Resources
Lipid Library
Edible Oil Processing
In the present context, the term edible oil processing covers the range of industrial…
Lipid Library
The Highs and Lows of Cannabis Testing
October 2016 With increasing legalization of both adult recreational and medical cannabis,…
Lipid Library
The secrets of Belgian chocolate
By Laura Cassiday May 2012 Like a bonbon nestled snugly in a…