1. Introduction
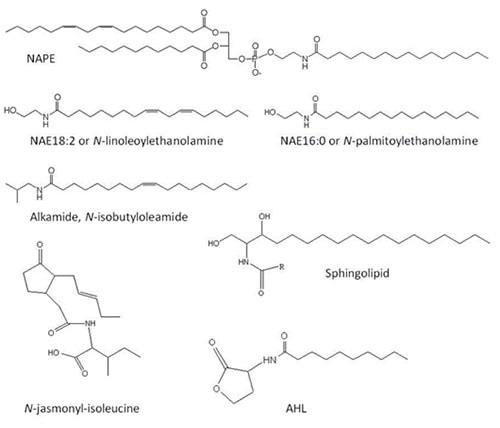
N-Acylphosphatidylethanolamines (NAPEs), N-acylethanolamines (NAEs) and other acylamides are nitrogen-containing lipids. Nitrogen-containing lipids are represented by a broad range of important structural and bioactive molecules in plant tissues including NAPEs, NAEs, alkamides, sphingolipids, and amino acid-conjugated fatty acids (e.g. N-jasmonyl-isoleucine) [1-5] (Fig. 1). The synthesis and functions of sphingolipids will be described in this web site by Edgar B. Cahoon. Here we will focus on the metabolism, occurrence and functions of other important acylamide compounds present in plant cells such as NAPE, NAE and alkamide.
Figure 1. Structure of different acylamides or nitrogen-containing lipids. NAPE, N-acylphosphatidylethanolamine; NAE, N-acylethanolamine; AHL, N-decanoyl-homoserine lactone.
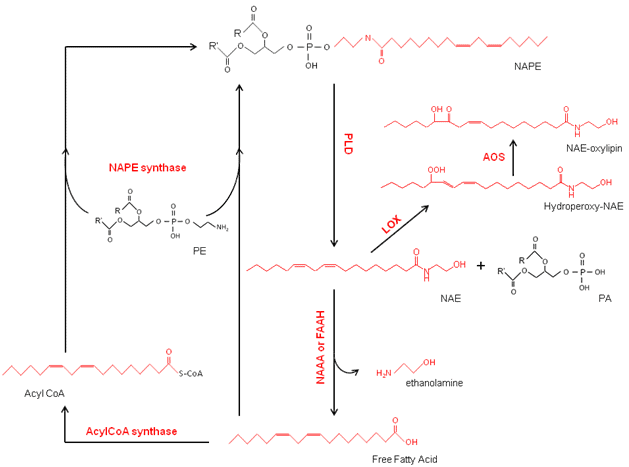
NAPE is generally considered to be present in all major groups of organisms including plants, animals, and some microbes. This class of glycerophospholipids is composed of phosphatidylethanolamine (PE) with a third fatty acid linked via an amide bond to the ethanolamine head group of the PE (Fig. 1). The molecular species compositions of NAPEs are complex with variations in the length and numbers of double bonds in the three acyl chains being described recently for Arabidopsis thaliana [6]. Described in detail below, in plants, NAPE is synthesized by N-acylation of PE by a NAPE synthase with either a free fatty acid (FFA) [7] or acyl-CoA substrate [8] (Fig. 2).
Figure 2. Metabolic pathway of NAPE/NAE formation in plants. NAPE, N-acylphosphatidylethanolamine; PLD, phospholipase D; NAE, N-acylethanolamine; LOX, lipoxygenase; AOS, allele oxide synthase; PA, phosphatidic acid; NAAA; N-acylethanolamine-hydrolyzing acid amidase; FAAH, fatty acid amide hydrolase; PE, phosphatidylethanolamine.
NAPEs can be used as precursors for the formation of NAEs by hydrolysis with one or more PLDs [9,10]. NAEs are found in virtually all types of eukaryotic organisms examined. Formation of NAEs from NAPEs has been intensively studied in mammalian systems as part of the so-called endocannabinoid signaling pathway, and several NAE types possess potent biological activities in human physiology and behavior [11-13]. The role of NAEs in plant systems is less well characterized, but the formation and turnover of NAEs appear to be associated with, among other stages, seed germination and seedling establishment (discussed in more detail below).
NAEs are metabolized in animal and plant systems by either hydrolysis or oxidation (Fig. 2). Hydrolysis of NAEs by fatty acid amide hydrolase (FAAH) to free fatty acid and ethanolamine has been characterized at the molecular level in both plants [14] and animals [15,16] whereas hydrolysis of NAEs by additional mechanisms has been described in animals [17-20]. Similarly, in both plants and animals, polyunsaturated NAEs can be oxidized by lipoxygenases (LOXs) to produce NAE-oxylipins [21-23]. In mammals, additional oxidative enzymes, most notably cyclooxygenases (COX), can act on polyunsaturated NAEs to produce ethanolamide oxylipins [24].
Contrary to NAEs (and NAPEs), the structurally related alkamides have been identified in only a few families of plants and some fungi [25]. The biosynthetic pathway of the alkamides and their functions are still not entirely clear. However, despite the similarity in structure to the NAEs, the formation of alkamides is not likely to proceed through the PLD-mediated hydrolysis of NAPEs.
2. NAPE Synthesis
The acylation of the PE with an acyl donor leads to the synthesis of NAPE. Several enzyme activities have been reported for the biosynthesis of NAPE, but only one activity has been identified at the molecular level. In animal cells, NAPE is reported to be synthesized by N-acylation of a phosphatidylethanolamine with the acyl group donor being a phospholipid, specifically the sn-1 position of a glycerophospholipid molecule (from either PE or PC) [26]. Three reactions have been described for this phospholipid-dependent acylation: (1) the Ca2+-dependent N-acyltransferase (NAT) [27], (2) the Ca2+-independent NAT [28] and (3) a phospholipase A/acyltransferase (PLA/AT) pathway [29]. For plants, two reactions have been described (Fig. 2), one that utilizes free fatty acids (FFA) as an acyl donor [7], and the other that uses acylCoA as an acyl donor [8]. An Arabidopsis gene, At1g78690, encoding an acyltransferase with the acylCoA-dependent NAPE synthase activity, was characterized recently.
2.1. The FFA- dependent N-acylation:
Although not identified at the molecular level, a 64 kDa membrane-bound enzyme able to transfer an acyl chain to the amine group of PE was partially purified from cottonseed by immobilized artificial membrane chromatography [7]. This enzyme uses the FFA as acyl donor for the direct acylation of PE by a reverse serine-hydrolase type catalytic mechanism. The enzyme exhibited complex cooperative kinetics toward free fatty acids, but more usual Michaelis-Menten type kinetics toward PEs. This enzyme activity was localized in the endoplasmic reticulum, Golgi and plasma membrane fractions isolated from cotyledons of cotton seedlings.
2.2. The AcylCoA-dependent N-acylation:
In 2010, a novel plasmalemmal protein was characterized as a NAPE synthase in vitro and in vivo in Arabidopsis thaliana. [8]. This protein belongs to the acyltransferase family and has three or four conserved regions with the PLsC acyltransferase motif characteristic of lysophosphatic acid acyltransferases (LPAATs) from E. coli and mammals [30]. The recombinant NAPE synthase protein uses acyl-CoA (16:0-CoA, 18:0-CoA) for the transfer of the acyl group to the amine group of PE. No significant activity was observed in vitro when the substrate used was a FFA. However, the function of this new protein is not entirely certain, since it was described to also have lysophosphatidylglycerol acyltransferase (LPGAT) activity, albeit only in vitro [31]. Thus, to date two different pathways have been described for the synthesis of NAPE in plants. However, a gene encoding a protein with FFA-dependent N-acylation activity remains to be identified. In terms of the acylCoA-dependent NAPE synthase, it is still not certain if this enzyme is sensu stricto a NAPE synthase, or if this acyltransferase protein also operates in planta for the synthesis of different glycerolipids such as phosphatidylglycerol (PG).
3. NAE Formation
In plants and in animals, the formation and turnover of NAE have been widely studied. In both systems, the acyl composition of the NAEs matches well with acyl compositions at the amide position of the NAPEs [3,6,32]. These compositional similarities along with radiolabeling studies [33,34] indicate that NAPE is the precursor for the NAE formation. In animals, a phospholipase D specific for NAPE substrate (NAPE-PLD) was cloned and well characterized [35]. This enzyme cleaves the terminal phosphodiester bond of NAPE to produce phosphatidic acid (PA) and NAE (Fig. 2). In addition, other alternative pathways for NAE formation from NAPE have been described [17-20]. These reactions involve different phospholipases acting on NAPE, such as (1) PLC-like followed by a phosphatase (PTPN22/SHIP1) that releases NAE, phosphate, and diacylglycerol, or (2) a deacylase (Abh4) releasing two fatty acids and glycerophospho-N-acyl-ethanolamine, followed by the action of glycerophosphoethanolamine diesterase (GDE1) releasing NAE and glycerol phosphate, or (3) PLA1/A2 releasing Lyso-NAPE and fatty acid, followed by the action of PLD-like release of NAE. In plants, the formation of NAEs is known to occur through different PLDs (below), but the precise PLD isoforms or additional mechanisms that might operate remain to be elucidated.
As in animals, the formation of NAEs in plants occurs via a phospholipase D-type (PLD) activity [35]. However, the PLD family in plants is composed of numerous isoforms (12 in Arabidopsis alone), and to date none of these PLDs have been strictly associated for the hydrolysis of NAPE to NAE. Among the 12 isoforms, only three (1 PLDα, 1PLDβ, 1PLDγ) have been tested for activity toward NAPE in vitro [10]. Further investigations are needed to assess if a functional ortholog of the animal NAPE-PLD exists in plants. The PLDα hydrolyses a variety of phospholipid substrates but is inactive toward the NAPE under conventional assay conditions in vitro. Actually, it has been demonstrated that the NAE is a negative regulator of the PLDα activity toward other phospholipid susbstrates [36]. The PLDβ and γ can both metabolize the NAPE to NAE in vitro and are so far considered to be two enzymes involved in the formation of NAEs in planta. Recently it was shown in Arabdiopsis that the NAE itself can modulate the content of NAE/NAPE via a negative feedback regulation [6]. Plants also may have additional pathways to form NAEs; however none have been described to date.
4. NAE Degradation
NAEs are characterized as lipid mediators in many different organisms. The regulation of the levels of NAEs, then, is a critical step in the NAE pathway in order to terminate biological effects in vivo. The metabolism of NAEs has been extensively studied [1-4]. In animals, the hydrolysis of NAE to FFA and ethanolamine is catalyzed by FAAH or by a N-acylethanolamine-hydrolyzing acid amidase (NAAA) [37] (Fig. 2). Alternatively, the polyunsaturated NAEs such as NAE18:2, NAE18:3, or NAE 20:4 can be oxygenated via LOX or COX to produce ethanolamide oxylipins with various potential bioactivities like vasomodulatory effects [38,39]. These compounds could have enhanced affinity with cannabinoid receptors in comparison with their respective nonoxygenated NAEs [40]. Alternatively, NAE-oxylipins such as prostaglandin ethanolamides may interact with novel receptors and modulate parameters such as intraocular blood pressure [41]. Similar pathways of hydrolysis or oxidation of NAEs also are found in plant cells.
4.1. Fatty acid amide hydrolase in plants
The fatty acid amide hydrolase belongs to the amidase super-family of proteins, and in Arabidopsis, there are seven proteins with the amidase signature sequence. So far in Arabidopsis, only one FAAH and one amidase (AMI-1) have been described that can hydrolyze NAE to FFA and ethanolamine (Fig. 2) [14,42]. Contrary to the animal FAAH, the reverse reaction (FFA + ethanolamine) was not catalyzed in plants [43]. On the other hand, Arabidopsis FAAH can hydrolyze a broad range of acyl amide and ester substrates in vitro, including the N-acylethanolamines, but does not appear to hydrolyze ceramide. A ser-ser-lys (S-S-K) catalytic triad-type mechanism is essential for the amido hydrolase activity of this protein [3]. The implication of FAAH in NAE metabolism in plants was confirmed by measuring the NAE content in seeds of plants with FAAH T-DNA insertions (knockouts) or in plants overexpressing the FAAH protein [44,45]. Since then, FAAH homologues in several plants have been characterized at the molecular and biochemical levels. Several additional FAAH candidates have been identified and still need to be characterized to better understand the regulation of NAE functions in plants. One other plant protein known to hydrolyze NAE in vitro is AMI-1. However, AMI-1 has only minor activity toward NAEs in vitro in comparison to its activity toward other compounds such as indole-3-acetamide and 1-naphthaleneacetamide [42].
4.2. NAE oxidation
A secondary pathway for NAE metabolism by LOX has been described in plant cells [44]. Contrary to the FAAH, the lipoxygenases don’t cleave the NAE, but instead oxidize the acyl chain of the polyunsaturated NAE (PU-NAE) like N-linoleoylethanolamine or (α,ϒ) N-linolenoylethanolamine to form NAE-oxylipins. The hydroperoxides of NAE compounds formed by the LOXes can be converted to additional NAE-oxylipins by hydroperoxide lyase (HPL) and allele oxide synthase (AOS) (Fig. 2) [21]. In vitro assays with 9-LOX from Solanum tuberosum (St), 13-LOX from Glycine max (Gm), or combined LOX activities in Arabidopsis homogenates, all exhibited substantial NAE-oxidation toward NAE18:2 or NAE18:3 [23]. In vivo, the oxidation of exogenous NAE18:2 or NAE18:3 has been also observed in young A. thaliana seedlings [23]. It appears that this LOX pathway can compete with FAAH in the overall metabolism of NAEs in plants [44].
5. Alkamide Metabolism
The alkamides are structurally similar to the NAEs (Fig. 1). They constitute a broad group of secondary metabolites (over 200) which have been found in 10 plant families [25]. The alkamides differ in the nature of their acyl groups, which typically are NOT similar to membrane fatty acids of the parent plant membranes, as well as the nature of the amide head group. Despite the large number of molecules comprising this class of lipid, no clear enzymatic pathway has been described for their synthesis in vivo. Because the “head groups” of the alkamides are propyl, isopropyl, butyl or isobutyl amides, as opposed to ethanolamides, it is unlikely that these compounds are formed from the PLD-mediated hydrolysis of NAPE. Instead it is likely that they have an alternative biosynthetic origin in the plant families in which they have been reported. Recent labeling experiments suggest that amino acids could be the precursor for the biosynthesis of the alkamides. Thus, valine or phenylalanine may be the precursors for the biosynthesis of some alkamides such as affinin ((2E,6Z,8E)-N-isobutyl-2,6,8-decatrienamide) [46]. Interestingly, FAAH may catalyze the hydrolysis of alkamides since this enzyme has been described as a good candidate for the hydrolysis of N-acyl-homoserine lactone (Fig. 1); this remains to be tested, however.
6. Occurrence of NAPEs/NAEs and Alkamides in Plants
In general, the amounts of NAPEs and NAEs in plant tissues are relatively minor. For example, NAPE is around 2-3% of total phospholipid content and this was relatively similar for different tissues and different growth stages of cotton seedlings and young plants [4,32]. NAPE and NAE content is usually highest in desiccated seeds [6], and for both lipid classes these levels declined with seedling establishment. In Arabidopsis, the level of NAEs in seeds was 2-2.6 µg/g, and this was reduced to 0.05 µg/g tissue fresh weight in seedling tissues [4,47]. Further, it has been reported that the NAE16C and 18C are predominant in dry seeds, while NAE12C and 14C are proportionately higher in the NAE pool of vegetative tissues [4]. In legume seeds, NAE profiles were similar to those in nonleguminous seeds, with concentrations ranging from 0.17 µg/g to 44.6 µg/g tissue fresh weight. The NAE acyl composition generally reflects the NAPE N-acyl composition, as well as the general membrane fatty acid compositions of plant tissues. However, the NAE and NAPE content and composition can change significantly with development or during different stresses such as the rehydration of desiccated seed [48], hypoxia (e.g., potato cells) [49] or pathogen elicitor perception [47].
Regarding alkamide distribution, it has been estimated that the highest concentration of affinin is up to 1% of fresh weight in the roots of Heliopsis longipes [50].
7. Amide Lipid Functions in Plants
Since the discovery of anandamide (NAE 20:4) as an endogenous ligand for the cannabinoid (CB) receptors in the mammalian nervous system [51], the studies of the functions of NAEs in animal systems have increased dramatically. In plants, the interest in NAEs and alkamides has been sparked by their pronounced effects on plant growth and development [52]. Work continues on the actions of acyl ethanolamides and alkamides in plants.
7.1. NAPE functions
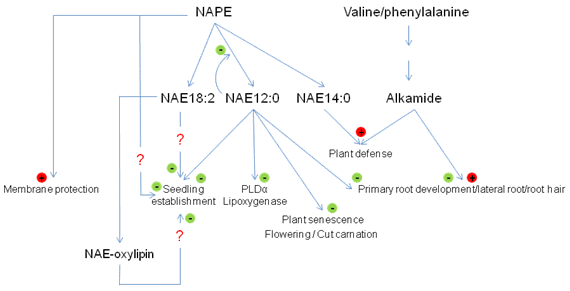
Biophysical studies with NAPE indicate that it has bilayer-stabilizing properties. Because it is synthesized from fatty acids and PE, both bilayer-destabilizing lipids, the synthesis of NAPE under certain stress conditions has been proposed to neutralize the “toxic effects” of FFA and PE and support the integrity and/or organization of the membrane. Therefore, the synthesis of NAPE under stress may help to keep the membranes functional [5,49]. NAPE biosynthesis has thus been described as a cytoprotective mechanism for membranes. Still, the main function attributed to this lipid in plants and animals is as the precursor of NAEs which carry out various lipid mediator functions in cells. On the other hand, NAPE functions may be more complex than previously appreciated, since the concentrations of these lipids also can be manipulated in conjunction with NAEs in Arabidopsis by altering the expression of FAAH [6].
7.2. NAE and alkamide functions
In animals, functions attributed to the NAE are numerous and continue to increase [1,53]. In plants the functions of NAE are also many and diverse. NAE12:0 is able to inhibit PLDα activity. The inhibition of PLDα by NAE12:0 may be partly involved in the regulation of stomata aperture [36]. It was also reported recently that NAE12:0 can competitively inhibit LOX activities, and so some physiological effects of NAE may be indirect though the modulation of oxylipin formation [22], as has been suggested for the NAE12:0-mediated delay in senescence of cut carnations (Dianthus caryophyllus) [54]. NAE metabolism may also participate in transition to flowering in Arabidopsis thaliana by modulating the transcription level of gene playing a crucial role for the flowering timing, the flowering locus T gene (FT) [55].
Figure 3. Some proposed functions of the NAPE, NAE and alkamide in plants. NAPE, N-acylphosphatidylethanolamine; NAE, N-acylethanolamine.
Addition of NAE12:0 to Arabidopsis seedlings results in a concentration-dependent disorganization of cell files and an inhibition of primary root growth, especially within the first few days of postgerminative growth. At the same time, exogenous NAE12:0 also inhibits the formation of the root hairs and the development of the lateral roots [4,52]. The arrest of early seedling growth by added NAEs and the normal, rapid depletion of endogenous NAEs during seedling establishment have prompted speculation that NAEs function as negative regulators of seedling growth. Indeed, an ABI 3-dependent interaction of NAE metabolism and seedling growth arrest was demonstrated in Arabidopsis indicating and intersection between NAE metabolism and ABA signaling in seedling development [56]. Other work suggests that NAEs like NAE14:0 may be involved in plant defense by modulating the expression of defense genes such as the phenylalanine ammonia lyase (PAL) or other early-membrane signaling events in plant pathogen interactions [47,57].
Alkamides share some apparent overlapping effects compared with NAE in plants. Exogenous N-isobutyl-decanamide (affinin) was shown to affect seedling growth, especially in root development. At low concentrations (7 µM), affinin induced an increase in the length of the primary roots, the emergence of lateral roots and the development of root hairs [25,58]. The increase of the lateral roots could be due to a stimulation of the emergence of pre-existing lateral root primordial, or via the synthesis of new ones. On the other hand, higher concentrations of affinin (120 µM) had the opposite effect on growth [25]. At high concentrations, this alkamide inhibited elongation of the primary root, the secondary root, and root hairs. Unlike the NAE, the decrease in root growth did not appear to be associated with a disruption of cell/tissue organization. Nevertheless, both types of lipids can negatively affect seedling development. Alkamides may interact with the ethylene pathway by altering the ethylene production or the signaling action of this hormone [25]. Affinin may also interact with the cytokinin-signaling pathway to control meristematic activity and cell differentiation events [58]. Also, like NAEs, alkamides affected plant defense responses against pathogens [59]. Affinin reduced the necrosis area induced by pathogens and inhibited fungal propagation. The mode of action of this alkamide for the plant defense is not fully understood. It might interact through the jasmonic acid pathway and MAP kinase-regulated signaling. Based on the structural similarity between alkamides and NAEs, it may be reasonable to speculate that some of the effects of alkamides on plants might be through endogenous targets of NAEs, since NAEs appear to be ubiquitous in the plant kingdom, and alkamides are more restricted in their distribution.
References
- Coulon, D., Faure, L., Salmon, M., Wattelet, V. and Bessoule, J. N-Acylethanolamines and related compounds: aspects of metabolism and functions. Plant Sci., 184, 129-140 (2012) (DOI: 10.1016/j.plantsci.2011.12.015).
- Kim, S., Chapman, K. and Blancaflor, E. Fatty acid amide lipid mediators in plants. Plant Sci., 178, 411-419 (2010) (DOI: 10.1016/j.plantsci.2010.02.017).
- Kilaru, A., Blancaflor, F., Venables, B., Tripathy, S., Mysore, K. and Chapman, K. The N-acylethanolamine-mediated regulatory pathway in plants. Chem. Biodivers., 4, 1933-1955 (2007) (DOI: 10.1002/cbdv.200790161).
- Chapman, K. Occurrence, metabolism, and prospective functions of N-acylethanolamines in plants. Prog. Lipid Res., 43, 302-327 (2004) (DOI: 10.1016/j.plipres.2004.03.002).
- Coulon, D., Faure, L., Salmon, M., Wattelet, V. and Bessoule, J. Occurrence, biosynthesis and functions of N-acylphosphatidylethanolamines (NAPE): Not just precursors of N-acylethanolamines (NAE). Biochimie, 94, 75-85 (2012) (DOI: 10.1016/j.biochi.2011.04.023).
- Kilaru, A., Tamura, P., Isaac, G., Welti, R., Venables, B.J., Seier, E. and Chapman, K.D. Lipidomic analysis of N-acylphosphatidylethanolamine molecular species in Arabidopsis suggests feedback regulation by N-acylethanolamines. Planta, 236, 809-824 (2012) (DOI: 10.1007/s00425-012-1669-z).
- McAndrew, R. and Chapman, K. Enzymology of cottonseed microsomal N-acylphosphatidylethanolamine synthase: Kinetic properties and mechanism-based inactivation. Biochim. Biophys. Acta, 1390, 21-36 (1998) (DOI: 10.1016/s0005-2760(97)00166-5).
- Faure, L., Coulon, D., Laroche-Traineau, J., Le Guedard, M., Schmitter, J., Testet, E., Lessire, R. and Bessoule, J. Discovery and characterization of an Arabidopsis thaliana N-acylphosphatidylethanolamine synthase. J. Biol. Chem., 284, 18734-18741 (2009) (DOI: 10.1074/jbc.M109.005744).
- Liscovitch, M., Czarny, M., Fiucci, G. and Tang, X. Phospholipase D: molecular and cell biology of a novel gene family. Biochem. J., 345, 401-415 (2000).
- Pappan, K., Austin-Brown, S., Chapman, K. and Wang, X. Substrate selectivities and lipid modulation of plant phospholipase Dα, -β, and -γ. Arch. Biochem. Biophys., 353, 131-140 (1998) (DOI: 10.1006/abbi.1998.0640).
- Lichtman, A.H., Varvel, S.A. and Martin, B.R. Endocannabinoids in cognition and dependence. Prostaglandins Leukotrienes Essent. Fatty Acids, 66, 269-285 (2002) (DOI: 10.1054/plef.2001.0351).
- Pacher, P., Bátkai, S. and Kunos, G. The endocannabinoid system as an emerging target of pharmacotherapy. Pharmacol. Rev., 58, 389-462 (2006) (DOI: 10.1124/pr.58.3.2).
- Maccarrone, M. Endocannabinoids: friends and foes of reproduction. Prog. Lipid Res., 48, 344-354 (2009) (DOI: 10.1016/j.plipres.2009.07.001).
- Shrestha, R., Dixon, R. and Chapman, K. Molecular identification of a functional homologue of the mammalian fatty acid amide hydrolase in Arabidopsis thaliana. J. Biol. Chem., 278, 34990-34997 (2003) (DOI: 10.1074/jbc.M305613200).
- Cravatt, B.F., Giang, D.K., Mayfield, S.P., Boger, D.L., Lerner, R.A. and Gilula, N.B. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature, 384, 83-87 (1996) (DOI: 10.1038/384083a0).
- Wei, B.Q., Mikkelsen, T.S., McKinney, M.K., Lander, E.S. and Cravatt, B.F. A second fatty acid amide hydrolase with variable distribution among placental mammals. J. Biol. Chem., 281, 36569-36578 (2006) (DOI: 10.1074/jbc.M606646200).
- Liu, J., Wang, L., Harvey-White, J., Osei-Hyiaman, D., Razdan, R., Gong, Q., Chan, A., Zhou, Z., Huang, B.X., Kim, H.-Y., and Kunos, G. A biosynthetic pathway for anandamide. Proc. Natl. Acad. Sci. U.S.A. 103, 13345-13350 (2006) (DOI: 10.1073/pnas.0601832103).
- Sun, Y., Tsuboi, K., Okamoto, Y., Tonai, T., Murakami, M., Kudo, I. and Ueda, N. Biosynthesis of anandamide and N-palmitoylethanolamine by sequential actions of phospholipase A2 and lysophospholipase D. Biochem. J., 380, 749-756 (2004) (DOI: 10.1042/bj20040031).
- Simon, G. and Cravatt, B. Endocannabinoid biosynthesis proceeding through glycerophospho-N-acyl ethanolamine and a role for α/β-hydrolase 4 in this pathway. J. Biol. Chem., 281, 26465-26472 (2006) (DOI: 10.1074/jbc.M604660200).
- Simon, G. and Cravatt, B. Anandamide biosynthesis catalyzed by the phosphodiesterase GDE1 and detection of glycerophospho-N-acyl ethanolamine precursors in mouse brain. J. Biol. Chem., 283, 9341-9349 (2008) (DOI: 10.1074/jbc.M707807200).
- Van Der Stelt, M., Noordermeer, M.A., Kiss, T., Van Zadelhoff, G., Merghart, B., Veldink, G.A. and Vliegenthart, J.F. Formation of a new class of oxylipins from N-acyl(ethanol)amines by the lipoxygenase pathway. Eur. J. Biochem., 267, 2000-2007 (2000) (DOI: 10.1046/j.1432-1327.2000.01203.x).
- Keereetaweep, J., Kilaru, A., Feussner, I., Venables, B. and Chapman, K. Lauroylethanolamide is a potent competitive inhibitor of lipoxygenase activity. FEBS Lett., 584, 3215-3222 (2010) (DOI: 10.1016/j.febslet.2010.06.008).
- Kilaru, A., Herrfurth, C., Keereetaweep, J., Hornung, E., Venables, B.J., Feussner, I. and Chapman, K.D. Lipoxygenase-mediated oxidation of polyunsaturated N-acylethanolamines in Arabidopsis. J. Biol. Chem., 286, 15205-15214 (2011) (DOI: 10.1074/jbc.M110.217588).
- Kozak, K.R. and Marnett, L.J. Oxidative metabolism of endocannabinoids. Prostaglandins Leukotrienes Essent. Fatty Acids, 66, 211-220 (2002) (DOI: 10.1054/plef.2001.0359).
- Ramírez-Chávez, E., López-Bucio, J., Herrera-Estrella, L. and Molina-Torres, J. Alkamides isolated from plants promote growth and alter root development in Arabidopsis. Plant Physiol., 134, 1058-1068 (2004) (DOI: 10.1104/pp.103.034553).
- Wang, J. and Ueda, N. Biology of endocannabinoid synthesis system. Prostaglandins Other Lipid Mediat., 89, 112-119 (2009) (DOI: 10.1016/j.prostaglandins.2008.12.002).
- Natarajan, V., Reddy, P.V., Schmid, P.C. and Schmid, H.H.O. N-Acylation of ethanolamine phospholipids in canine myocardium. Biochim. Biophys. Acta, 712, 342-355 (1982) (DOI: 10.1016/0005-2760(82)90352-6).
- Jin, X.H., Okamoto, Y., Morishita, J., Tsuboi, K., Tonai, T. and Ueda, N. Discovery and characterization of a Ca2+-independent phosphatidylethanolamine N-acyltransferase generating the anandamide precursor and its congeners. J. Biol. Chem., 282, 3614-3623 (2007) (DOI: 10.1074/jbc.M606369200).
- Uyama, T., Ikematsu, N., Inoue, M., Shinohara, N., Jin, X.H., Tsuboi, K., Tonai, T., Tokumura, A. and Ueda, N. Generation of N-acylphosphatidylethanolamine by members of the phospholipase A/acyltransferase (PLA/AT) family. J. Biol. Chem., 287, 31905-31919 (2012) (DOI: 10.1074/jbc.M112.368712).
- Lewin, T.M., Wang, P. and Coleman, R.A. Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry, 38, 5764-5771 (1999) (DOI: 10.1021/bi982805d).
- Bulat, E. and Garrett, T.A. Putative N-acylphosphatidylethanolamine synthase from Arabidopsis thaliana is a lysoglycerophospholipid acyltransferase. J. Biol. Chem., 286, 33819-33831 (2011) (DOI: 10.1074/jbc.M111.269779).
- Chapman, K., Venables, B., Markovic, R., Blair, R. and Bettinger, C. N-Acylethanolamines in seeds. Quantification of molecular species and their degradation upon imbibition. Plant Physiol., 120, 1157-1164 (1999) (DOI: 10.1104/pp.120.4.1157).
- Schmid, H.H., Schmid, P.C. and Natarajan, V. N-Acylated glycerophospholipids and their derivatives. Prog. Lipid Res., 29, 1-43 (1990).
- Chapman, K., Lin, I. and Desouza, A. Metabolism of cottonseed microsomal N-acylphosphatidylethanolamine. Arch. Biochem. Biophys., 318, 401-407 (1995) (DOI: 10.1006/abbi.1995.1246).
- Okamoto, Y., Morishita, J., Tsuboi, K., Tonai, T. and Ueda, N. Molecular characterization of a phospholipase D generating anandamide and its congeners. J. Biol. Chem., 279, 5298-5305 (2004) (DOI: 10.1074/jbc.M306642200).
- Austin-Brown, S. and Chapman, K. Inhibition of phospholipase Dα by N-acylethanolamines. Plant Physiol., 129, 1892-1898 (2002) (DOI: 10.1104/pp.001974).
- Ueda, N., Tsuboi, K. and Uyama, T. N-Acylethanolamine metabolism with special reference to N-acylethanolamine-hydrolyzing acid amidase (NAAA). Prog. Lipid Res., 49, 299-315 (2010) (DOI: 10.1016/j.plipres.2010.02.003).
- Pratt, P.F., Hillard, C.J., Edgemond, W.S. and Campbell, W.B. N-Arachidonylethanolamide relaxation of bovine coronary artery is not mediated by CB1 cannabinoid receptor. Am. J. Physiol., 274, H375-381 (1998).
- Kunos, G., Járai, Z., Bátkai, S., Goparaju, S.K., Ishac, E.J., Liu, J., Wang, L. and Wagner, J.A. Endocannabinoids as cardiovascular modulators. Chem. Phys. Lipids, 108, 159-168 (2000).
- Hampson, A.J., Hill, W.A., Zan-Phillips, M., Makriyannis, A., Leung, E., Eglen, R.M. and Bornheim, L.M. Anandamide hydroxylation by brain lipoxygenase: metabolite structures and potencies at the cannabinoid receptor. Biochim Biophys Acta, 1259, 173-179 (1995).
- Woodward, D.F., Krauss, A.H., Chen, J., Lai, R.K., Spada, C.S., Burk, R.M., Andrews, S.W., Shi, L., Liang, Y., Kedzie, K.M., et al. The pharmacology of bimatoprost (Lumigan). Surv. Ophthalmol., 45, S337-345 (2001).
- Pollmann, S., Neu, D., Lehmann, T., Berkowitz, O., Schafer, T. and Weiler, E. Subcellular localization and tissue specific expression of amidase 1 from Arabidopsis thaliana. Planta, 224, 1241-1253 (2006) (DOI: 10.1007/s00425-006-0304-2).
- Shrestha, R., Kim, S., Dyer, J., Dixon, R. and Chapman, K. Plant fatty acid (ethanol) amide hydrolases. Biochim. Biophys. Acta, 1761, 324-334 (2006) (DOI: 10.1016/j.bbalip.2006.03.004).
- Shrestha, R., Noordermeer, M., Van der Stelt, M., Veldink, G. and Chapman, K. N-Acylethanolamines are metabolized by lipoxygenase and amidohydrolase in competing pathways during cottonseed inbibition. Plant Physiol., 130, 391-401 (2002) (DOI: 10.1104/pp.004689).
- Wang, Y., Shrestha, R., Kilaru, A., Wiant, W., Venables, B., Chapman, K. and Blancaflor, E. Manipulation of Arabidopsis fatty acid amide hydrolase expression modifies plant growth and sensitivity to N-acylethanolamines. Proc. Natl. Acad. Sci. U.S.A.,, 103, 12197-12202 (2006) (DOI: 10.1073/pnas.0603571103).
- Cortez-Espinosa, N., Avina-Verduzco, J., Ramirez-Chavez, E., Molina-Torres, J. and Rios-Chavez, P. Valine and phenylalanine as precursors in the biosynthesis of alkamides in Acmella radicans. Nat. Prod. Commun., 6, 857-861 (2011).
- Tripathy, S., Venables, B. and Chapman, K. N-Acylethanolamines in signal transduction of elicitor perception. Attenuation of alkalinization response and activation of defense gene expression. Plant Physiol., 121, 1299-1308 (1999) (DOI: 10.1104/pp.121.4.1299).
- Chapman, K.D. and Moore, T.S. Catalytic properties of a newly discovered acyltransferase that synthesizes N-acylphosphatidylethanolamine in cottonseed (Gossypium hirsutum L.) microsomes. Plant Physiol., 102, 761-769 (1993).
- Rawyler, A.J. and Braendle, R.A. N-Acylphosphatidylethanolamine accumulation in potato cells upon energy shortage caused by anoxia or respiratory inhibitors. Plant Physiol., 127, 240-251 (2001).
- López-Bucio, J., Acevedo-Hernández, G., Ramírez-Chávez, E., Molina-Torres, J. and Herrera-Estrella, L. Novel signals for plant development. Curr. Opin. Plant. Biol., 9, 523-529 (2006) (DOI: 10.1016/j.pbi.2006.07.002).
- Devane, W.A., Hanus, L., Breuer, A., Pertwee, R.G., Stevenson, L.A., Griffin, G., Gibson, D., Mandelbaum, A., Etinger, A. and Mechoulam, R. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science, 258, 1946-1949 (1992).
- Blancaflor, E., Hou, G. and Chapman, K. Elevated levels of N-lauroylethanolamine, an endogenous constituent of desiccated seeds, disrupt normal root development in Arabidopsis thaliana seedlings. Planta, 217, 206-217 (2003) (DOI: 10.1007/s00425-003-0985-8).
- Ueda, N., Tsuboi, K. and Uyama, T. Enzymological studies on the biosynthesis of N-acylethanolamines. Biochim. Biophys. Acta, 1801, 1274-1285 (2010) (DOI: 10.1016/j.bbalip.2010.08.010).
- Zhang, Y., Guo, W.M., Chen, S.M., Han, L. and Li, Z.M. The role of N-lauroylethanolamine in the regulation of senescence of cut carnations (Dianthus caryophyllus). J. Plant. Physiol., 164, 993-1001 (2007) (DOI: 10.1016/j.jplph.2006.07.003).
- Teaster, N.D., Keereetaweep, J., Kilaru, A., Wang, Y.S., Tang, Y., Tran, C.N., Ayre, B.G., Chapman, K.D. and Blancaflor, E.B. Overexpression of fatty acid amide hydrolase induces early flowering in Arabidopsis thaliana. Front. Plant Sci., 3, 32 (2012) (DOI: 10.3389/fpls.2012.00032).
- Cotter, M.Q., Teaster, N.D., Blancaflor, E.B. and Chapman, K.D. N-Acylethanolamine (NAE) inhibits growth in Arabidopsis thaliana seedlings via ABI3-dependent and -independent pathways. Plant Signal. Behav., 6, 671-679 (2011).
- Tripathy, S. and Chapman, K. Regulation of N-acylphosphatidylethanolamine biosynthesis in elicitor-treated tobacco cells. Plant Physiol., 111, 704-704 (1996).
- López-Bucio, J., Millán-Godínez, M., Méndez-Bravo, A., Morquecho-Contreras, A., Ramírez-Chávez, E., Molina-Torres, J., Pérez-Torres, A., Higuchi, M., Kakimoto, T. and Herrera-Estrella, L. Cytokinin receptors are involved in alkamide regulation of root and shoot development in Arabidopsis. Plant Physiol., 145, 1703-1713 (2007) (DOI: 10.1104/pp.107.107953).
- Méndez-Bravo, A., Calderón-Vázquez, C., Ibarra-Laclette, E., Raya-González, J., Ramírez-Chávez, E., Molina-Torres, J., Guevara-García, A.A., López-Bucio, J. and Herrera-Estrella, L. Alkamides activate jasmonic acid biosynthesis and signaling pathways and confer resistance to Botrytis cinerea in Arabidopsis thaliana. PLoS One, 6, e27251 (2011) (DOI: 10.1371/journal.pone.0027251).
Related Resources
Lipid Library
Edible Oil Processing
In the present context, the term edible oil processing covers the range of industrial…
Lipid Library
The Highs and Lows of Cannabis Testing
October 2016 With increasing legalization of both adult recreational and medical cannabis,…
Lipid Library
The secrets of Belgian chocolate
By Laura Cassiday May 2012 Like a bonbon nestled snugly in a…