Introduction
Phospholipases are enzymes that occur ubiquitously across all kingdoms of life and catalyze the cleavage of phospholipid molecules. Phospholipids play an important role as the main structural elements of cellular membranes but they, together with their cleavage products that arise from the action of the different phospholipases, also fulfill several key regulatory functions in the cell. The main areas of action of phospholipases are the participation in remodeling of cellular membranes, membrane trafficking and lipid-mediated signal transduction processes, cell proliferation, and apoptosis. In microorganisms, phospholipases play additionally important roles in the nutrient acquisition, cell-cell communication, virulence, and penetration of the host.
Microbial phospholipases, mainly from Gram-negative bacteria, are involved in several pathogenic processes linked to various diseases. In these roles they act either directly by causing severe cell damage and consequent destruction of membranes or by interfering with signal transduction processes within the host or by helping to evade the host organism’s immune defense mechanisms. Sequencing of numerous microbial genomes has led to the identification of a multitude of new genes whose products are predicted to act on phospholipids. However, many of the proteins encoded by these genes are still awaiting a correct biochemical analysis for an unambiguous classification.
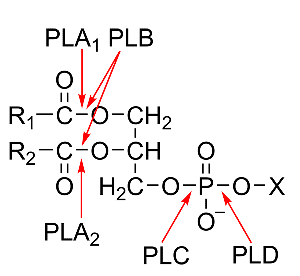
The classification of phospholipases is based on their site of attack on the phospholipid molecule and is shown in Fig. 1. The acyl hydrolases phospholipases A1 (PLA1) and A2 (PLA2) cleave the acyl ester bonds in sn-1 and sn-2 position, respectively, whereas phospholipase B (PLB) does not show a preference for either of these positions. The phosphodiesterase, phospholipase C (PLC), catalyzes the cleavage of the glycerophosphate bond, whereas phospholipase D (PLD) cleaves the other phosphodiester bond in the head group of the phospholipid molecule.
Figure 1: Structure of phospholipids and sites of attack by the different phospholipases. The non-polar chains of the fatty acids esterified to the glycerol backbone in the sn-1 and sn-2 positions are presented as R1 and R2, the alcohol esterified to the phosphoryl residue in sn-3 position is depicted as X.
1. Microbial PLAs
Due to the often very complex pattern of activities displayed by those acyl hydrolases and the lack of biochemical data, their nomenclature [with the exception of the secreted PLA2s (sPLA2)] poses a significant problem. To classify new microbial acyl hydrolases unambiguously, and to exclude the interference by competing downstream activities, the use of purified enzymes and reliable and reproducible activity assays is crucial.
1.1. Microbial PLA1
Principally, PLA1s are enzymes that hydrolyze glycerophospholipids at the acyl ester bond in sn-1 position and produce a 2-acyl-lysophospholipid and a free fatty acid (Fig. 1). The unambiguous assignment of enzymes to this group (EC 3.1.1.32) and the separation from PLBs and esterases/lipases proves to be difficult, especially for the enzymes from fungi, due to their broad substrate specificity. Most of the PLA1s classified so far are in fact PLB-type enzymes. In addition, microbial PLA1s could also be considered as members of the esterase-lipase superfamily because their sequences strongly resemble some hepatic, pancreatic and lipoprotein lipases from animals, which very often also display significant PLA1 activity. An example is the lipase from Streptomyces hyicus. Therefore, one criterion for unambiguous classification might be the inability to act on triacylglycerols.
All of these enzymes possess the G-X-S-X-G lipase consensus motif. Catalysis proceeds by the formation of an acyl enzyme intermediate of the phospholipid with the Ser residue, which is part of the canonical catalytic His-Ser-Asp triad, of this motif. Their molecular mass lies between 30 – 40 kDa. Despite the fact that no crystal structure of any true PLA1 is available, sequence alignments suggest the presence of the common α/β-hydrolase fold corresponding to the fold inherent in the other members of this superfamily. The enzymes do not require divalent metal ions for activity but are stimulated by Mg2+ and Ca2+.
PLA1s are found in several fungi (Aspergillus, Fusarium, Serratia, Thermomyces, Bacterolides strains), protozoa (Tetrahymena, Trichomonas, trypanosoma strains), Gram-positive (Corynebacteria, Nocardia strains) and especially Gram-negative bacteria (Serratia, Yersinia, Pseudomonas, Lysobacter, Aeromonas, Enterobacter, Xenorhabdus, Photorhabdus, Xanthomonas strains). Some of them play potential roles as virulence factors; others might exert their function by the action of the lysophospholipid product.
A special case is represented by the outer membrane phospholipase A (OMPLA) (EC 3.1.1.4 and 3.1.1.32) – an integral membrane protein of many Gram-negative bacteria such as E. coli, Klebsiella, Salmonella, Enterobacter, Helicobacter, Citrobacter, Serratia, and Proteus strains. Even though its activity towards the acyl ester bond in the sn-1 position is 6-fold higher than that towards the acyl ester bond in sn-2 position, it is more like a PLB than a PLA1,as usually citedin the literature. In addition, OMPLA shows lysophospholipase activity and, like PLBs, also mono- and diacylglycerol lipase activity. OMPLA is involved in bacteriocin secretion. The amino acid sequence is extremely well conserved across all species (95-99 %). The best studied and crystallized OMPLA from E. coli has a molecular mass of 31 kDa. Binding of Ca2+ promotes the reversible formation of a dimeric structure with the Ca2+ at the dimer interface, which is essential for expression of activity. The atypical Ser-His-Asn catalytic triad is located at the outer leaflet side of the membrane.
1.2. Microbial PLA2
PLA2s (EC 3.1.1.4) stereospecifically catalyze the hydrolysis of the acyl ester bond of phospholipids in the sn-2 position of the glycerol backbone to release the corresponding lysophospholipid and a free fatty acid. The products of the reaction have many important physiological functions. In general, the large superfamily of PLA2s is subdivided into several classes – secreted PLA2s (sPLA2), cytosolic (cPLA2), Ca2+-independent (iPLA2), platelet-activating factor-hydrolyzing (PAF-AH-PLA2), lysosomal (lPLA2), and the recently described adipose-specific PLA2s. Categorization into groups I to XVI depends on molecular characteristics, catalytic mechanism (using a His/Asp, Ser/Asp or Ser/His/Asp dyad or triad, respectively), functional and structural features, domain organization, localization, Ca2+ ion requirement, and number of disulfide bonds.
In microorganisms, three types of low molecular weight sPLA2 as well as patatin- and cPLA2-like PLA2 have been found.
1.2.1. Microbial sPLA2
1.2.1.1. Prokaryotic sPLA2
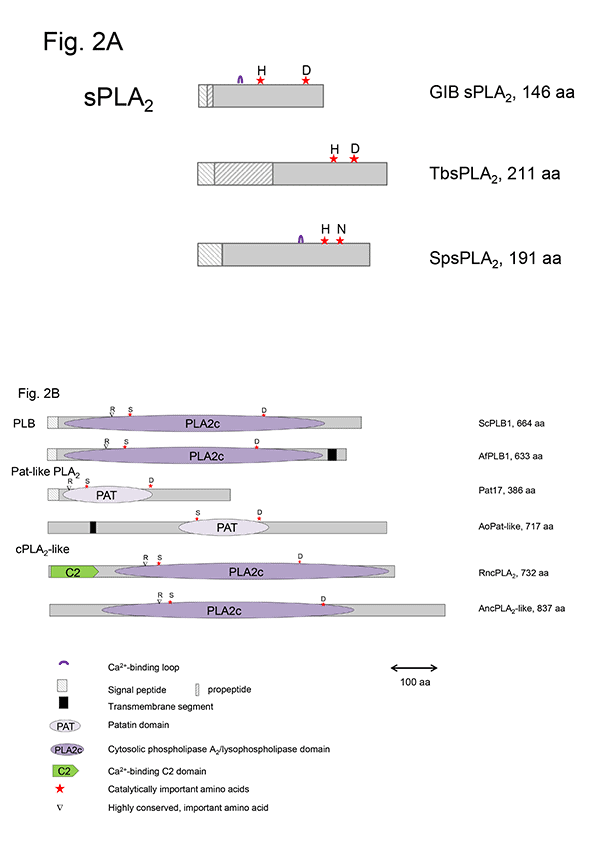
This group of sPLA2s (GXIV, prokaryotic PLA2) contains the conserved domain pfam09056. Besides the well-characterized enzymes from Streptomyces violaceoruber and Strept. coelicolor and the ascomycete Tuber borchii, related enzymes are predicted to exist in various Gram-positive (Actinoplanes, Deinococcus, Nocardia strains) and Gram-negative bacteria (Mitsuaria, Thauera strains), as well as several uni- and multicellular fungi (Helicosporium, Aspergillus, Fusarium, Trichoderma, Magnaporthe, Neurospora, Coccioides strains). The crystal structures of the enzymes from Strept. violaceoruber (1FAZ) and T. borchii (4AUP) show that they are mainly composed of five α-helices and are different to those of most eukaryotic sPLA2s. The catalytic His/Asp dyad is well-conserved in all members of this group; however, there is no Ca2+-binding loop typical of most eukaryotic sPLA2s (Fig. 2A). Ca2+-binding is enabled via the Asp residue adjacent to the catalytic His and by Gly or Leu and Asp residues upstream of the catalytic site. In contrast to most other sPLA2s, these enzymes contain only four conserved Cys residues forming two disulfide bonds.
Figure 2: Schematic presentation of microbial sPLA2, PLB, cPLA2- and patatin-like PLA2 in comparison to representative mammalian and plant enzymes.Red stars highlight the catalytically active amino acids, and ▼ indicates one highly conserved important amino acid. A. Microbial sPLA2: GIB – bovine pancreas sPLA2, TbsPLA2 – GXIV – T. borchii sPLA2 (Pfam09056), SpsPLA2 – S. pyogenes sPLA2 (cd04706) B. Microbial PLB, patatin- and cPLA2-like PLA2: ScPLB1 – PLB1 (S. cerevisiae), AfPLBA – PLB1 (A. fumigatus), Pat17 – patatin17 (Solanum cardiophyllum), AoPat-like -Patatin-like phospholipase (A. oryzae), RncPLA2 – cPLA2 (Rattus norvegicus), AncPLA2-like – cPLA2-like phospholipase (A. nidulans)
1.2.1.2. Plant-related sPLA2s
In different Gram–positive and Gram–negative bacteria, such as Streptococcus pyogenes, Clostridium perfringens and Bacillus cereus, sPLA2s with the conserved domain cd04706 (plant-specific PLA2) have been identified. These enzymes share a number of conserved structural elements with sPLA2s from plants (GXI) such as the catalytic His residue, the Ca2+-binding loop, substitution of the highly conserved Asp residue of the catalytic dyad by Asn, His or Ser residues, and two conserved disulfide bonds (Fig. 2A). However, these microbial enzymes await a more detailed biochemical characterization.
1.2.1.3. Viral sPLA2s
sPLA2 from almost all parvoviruses (GXIII) are encoded by a relatively short sequence stretch in the N-terminal part of the virus capsid protein VP1. They differ from the other sPLA2s in structure and catalytic activity but contain the canonical His/Asp dyad and a conserved Ca2+-binding loop. The enzyme possesses no disulfide bonds and was shown to play a role in infectivity of the virus.
1.2.2. Patatin-like microbial PLA2s
The opportunistic pathogenic Gram–negative bacterium, Pseudomonas aeruginosa, produces the potent cytotoxin ExoU (74 kDa), which is responsible for targeting of the host cell via a type III secretion system and is activated by ubiquitin. ExoU, as well as VipD from Legionella pneumophila,contain patatin-like domains. BLAST searches revealed the existence of similar enzymes in many other bacteria (Burkholderia, Aeromonas, B. subtilis, Rickettsia strains), yeasts (S. cerevisiae), and fungi (such as Aspergillus, Colletotrichum, Candida and Coccioides strains) too. Bioinformatic analyses showed that the patatin-like microbial enzymes closely resemble iPLA2s or patatin-like PLA2s from animals and plants (Fig. 3). Patatins are unspecific acyl hydrolases that hydrolyze phospholipids, glycolipids, sulfolipids, mono– and diacylglycerols.
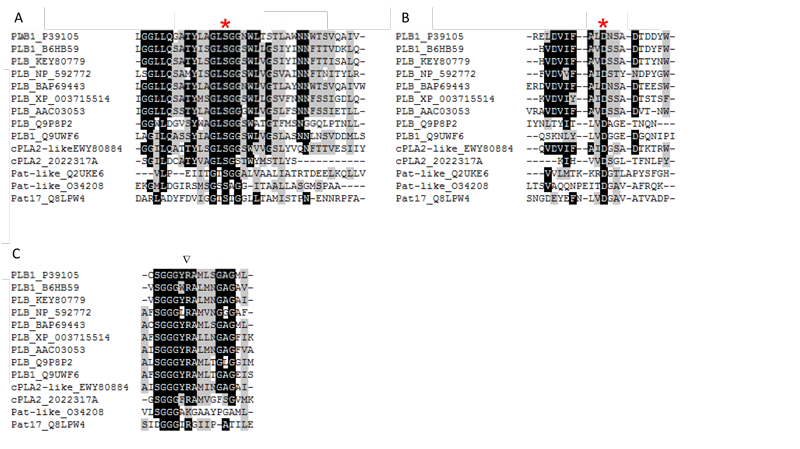
Figure 3: Conserved regions around catalytically important amino acids in the active site of microbial PLBs, cPLA2- and patatin-like domains containing microbial enzymes. B6HB59 (PLB1 P. chrysogenum), P39105 (PLB1 S. cerevisiae), KEY80779 (PLB A. fumigatus), NP_592772 (predicted PLB S. pombe), BAP69443 (PLB K. marxianus), XP_003715514 (PLB Magnaporthe oryzae), AAC03053 (PLB N. crassa), Q9P8P2 (PLB C. neoformans), Q9UWF6 (PLB1 C. albicans), Q2UKE6 (patatin-like PLA A. oryzae), EWY80884 (cPLA2-like PLA2 F. oxysporum), 2022317A (cPLA2 R. norvegicus), O34208 (patatin-like PLA ExoU P. aeruginosa), Q8LPW4 (Patatin 17 S. cardiophyllum).
In contrast to the sPLA2s, the microbial enzymes that contain patatin-like domains (pfam 01734) utilize a conserved active site dyad composed of a nucleophilic Ser residue, located in the conserved lipase consensus G-X-S-X-G motif, and an Asp residue. In addition, a conserved Arg residue is required for activity in patatin- and cPLA2-like PLA2s as well as PLB (Fig. 2B, 3). The patatins possess an α,β-hydrolase fold; however, they do not contain a lid domain and are not subjected to interfacial activation. In contrast to sPLA2s strictly acting only on the acyl ester bond in the sn-2 position of phospholipids, the microbial patatin-like PLA2s also display PLA1 and lysophospholipase activities and have broad substrate specificity cleaving not only phospholipids. Due to the lack of crystal structures, no structural and mechanistic explanations for the multiple enzyme activities displayed by these enzymes can be provided.
1.2.3. Microbial cPLA2-like enzymes
Genes encoding cPLA2-like proteins, which resemble human GIV cPLA2s, have been identified in the genomes of several fungi, including Aspergillus (especially PLaA of A. nidulans), Neurospora, Colletotrichum, and Fusarium strains but most of the encoded proteins need better biochemical characterization. The lack of signal peptides points to an intracellular localization in the fungi. Catalysis is fulfilled by similar amino acids and motifs described above for patatin-like PLA2s (Fig. 2B, 3). The fungal cPLA2-like enzymes do not contain the C2 domain which is an essential element of mammalian cPLA2s (Fig. 2B). However, their sequences have a higher similarity to mammalian cPLA2s rather than fungal PLBs as indicated by BLAST searches. cPLA2s require µM concentrations of Ca2+ for membrane targeting via the C2 domain rather than for catalysis, whereas towever, he known fungal cPLA2-like enzyme requires Ca2+ for expression of activity. The main function of mammalian cPLA2s is the release of arachidonic acid from the sn-2 position of phospholipids as the precursor of eicosanoids. Whether the cPLA2-like enzymes from fungi might play a similar role in production of oxylipin precursors can only be speculated.
1.2.4. Microbial PLB
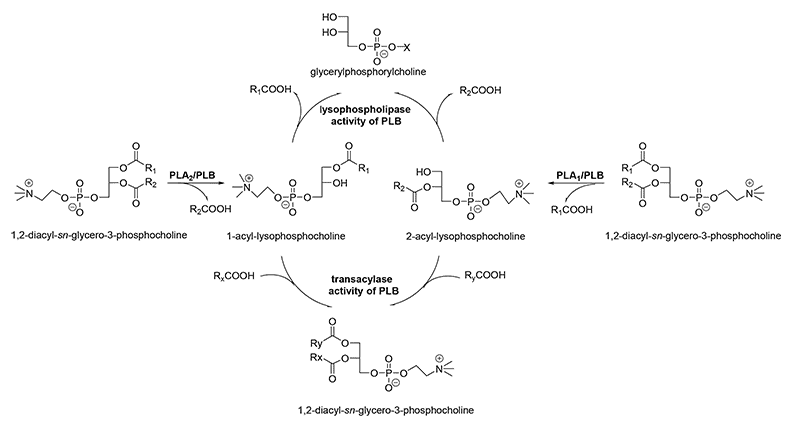
Many fungal species (Aspergillus, Penicillium, Neurospora, Cryptococcus strains) and yeasts (Saccharomyces cerevisiae, Candida, Schizosaccharomyces, Torulaspora, Klyuveromyces strains) produce one or more enzymes that can cleave both acyl ester bonds of glycerophospholipids to release free fatty acids and the corresponding lysophospholipids. The subsequent release of acyl chains from both types of the primarily produced lysophospholipids is achieved by an inherent lysophospholipase activity that results in formation of glycerophosphodiesters (Fig. 4). Thus, minimal amounts of lysophospholipids are accumulated.
Figure 4: Reactions catalyzed by PLA1, PLA2, and PLB.
In addition, these enzymes possess a transacylase activity, which leads to incorporation of fatty acids into lysophospholipids (Fig. 4). These features altogether lead to the designation as PLBs, and the concomitant cleavage of phospholipids at the sn-1 and sn-2 positions alone is not sufficient. Most of PLBs from pathogenic microorganisms prefer PC, whereas the enzymes from non-pathogenic microorganisms are described to prefer PS, PI, and PA being mostly located at the inner leaflet of the plasma membrane. The molecular masses of PLBs are in the range of 65 to 90 kDa. Some PLBs, especially those from yeast and several parasites, are characterized by exhibiting extensive glycosylation and having a GPI anchor. The best studied enzyme of this class is the PLB from Cryptococcus neoformans. Sequence alignments and mutational studies showed that catalysis is performed via a highly conserved Ser/Asp dyad with a Ser residue in the canonical GxSxG motif (Fig. 2B). The Ser residue functions as a nucleophile in the reaction to form an enzyme-acyl intermediate. The Asp residue acts as general base and activates the catalytic Ser residue. An essential and highly conserved Arg residue is not directly involved in catalysis but might stabilize the phospholipid head group. All of the different catalytic activities mentioned above are performed by one active site with potentially two binding pockets. Homology modeling showed that the core domain with the active site resembles the comparable structural region of mammalian cPLA2s legitimizing the classification into cd07203 (cPLA2_Fungal_PLB).
2. Microbial lysophospholipases
The distinction between microbial PLBs and lysophospholipases and the definition of true lysophospholipases is a very complicated issue as most of the PLBs also possess some lysophospholipase activity. In general, this class of enzymes is not well characterized and many microbial enzymes probably have incorrectly been annotated because some of them additionally possess mainly esterase activity but can also act on 1,2-diacyl-sn-glycero-3-phospholipids. Lysophospholipase activity plays an important role in controlling the level of lysophospholipids in the cell, thereby contributing to membrane stabilization. Mostly, lysophospholipases can act equally well on acyl ester bonds in sn-1 and sn-2 position. An important feature of lysophospholipases is the inhibition by detergents in contrast to phospholipases A and B which prefer mixed micelles of phospholipids and detergents as substrates.
3. Microbial PLCs
PLCs are widely distributed in Gram-negative and Gram-positive bacteria where they mostly play important roles in pathogenesis and host cell invasion and cytotoxicity. They cleave the glycero-phosphodiester bond of phospholipids as depicted in Fig. 1. In their dependence on the preferred substrate, three main classes of PLCs are known. They comprise PC-PLCs, primarily acting on PC (phosphatidyl choline) or PC/sphingomyelin (EC 3.1.4.3, Fig. 5A), PI-PLCs, cleaving phosphatidyl inositol (PI) or the glycosyl phosphatidyl inositol (GPI) anchors of mammalian proteins, but not inositol phosphates (EC 4.6.1.13, Fig. 5B), because the active site cleft of these PLCs is not large enough to accommodate phosphoinositides with more than one phosphate group, and PLCs that favour phosphatidyl inositol phosphates as substrates (EC 3.1.4.11, Fig. 5C). A poorly described GPI-specific PLC (EC 4.6.1.14, Fig. 5D) is expressed in Trypanosoma brucei.
Figure 5: Reactions catalyzed by different classes of PLC.
A: PC-specific PLC (EC 3.1.4.3)
B: Phosphatidylinositol-specific PLC (EC 4.6.1.13)
C: Phosphoinositide-specific PLC (EC 3.1.4.11)
D: Glycosylphosphatidylinositol-specific PLC (EC 4.6.1.14)
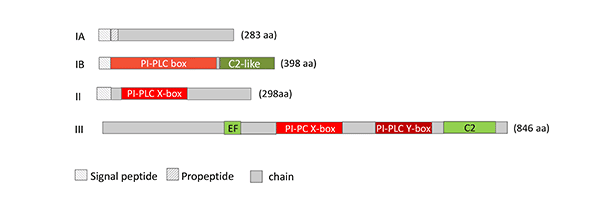
Members of PC-PLCs and PI-PLCs are found in different Bacillus and Clostridium strains, Staphylococcus aureus, Burkholderia pseudomallei, Legionella pneumophila, diverse Mycobacteria, Plasmodium falciparum, Pseudomonas strains, especially P. aeruginosa, Pseudoalteromonas sp., Tetrahymena pyriformis. The molecular masses of Group 1 PC-PLCs and PI-PLCs are in the range of 28 – 34 kDa. Group 2 PC-PLCs are mainly found in Gram-negative bacteria, such as Pseudomonas aeruginosa, are larger (about 70 – 80 kDa). The 800 – 1000 amino acids comprising phosphoinositide-specific PLCs are mainly found in yeasts, such as S. cerevisiae, Klyuveromyces and Candida strains, and in slime molds (Fig. 6).
Figure 6: Schematic representation of the structure of microbial PLC. IA: Zinc-dependent, PC-specific PLC (EC 3.1.4.3; P09598, B. cereus), IB: zinc-dependent, PC-specific PLC (EC 3.1.4.3; BAA09944, C. perfringens), II: phosphatidylinositol-specific PLC (EC 4.6.1.13; 1PTD_A, B. cereus), III: phosphoinositide-specific PLC from S. cerevisiae (EC 3.1.4.11; P32383).
The best studied microbial PLCs are the secreted PC-PLCs from Bacillus cereus, the α-toxin from Clostridium perfringens, the PI-PLCs from B. cereus, Staphylococcus aureus, Streptomyces antibioticus, and Listeria monocytogenes. Their crystal structures (1AH7, 1CA1, 1PTD, 3V16, 3H4W, 2PLC, respectively) allowed the elucidation of mechanistic details for these two classes of enzymes. In all cases, catalysis is based on a general acid/base mechanism. PLCs of a different type and origin can display different types of amino acid residues that fulfill the role of the general acid and base in catalysis. In most cases, His residues are predominant and are coupled to an Asp residue in a charge-relay system to enhance the nucleophilic power of the catalytic His residue as general base. Three essential Zn ions play an important role in catalysis only of the PC-PLCs of Group 1. Conversion of substrates by microbial PI-PLCs (EC 4.6.1.13), and probably of the less well-characterized phosphoinositide-specific yeast enzymes of class EC 3.1.4.11, is a two-step process. In the first step, a phosphotransferase reaction, the activated C2-hydroxyl group of the inositol moiety intra-molecularly attacks the adjacent phosphate group to form myo-inositol 1,2-cyclic phosphate or cyclic inositoltrisphosphate (IP3), respectively, as stable intermediates. Mammalian enzymes and microbial enzymes of class EC 3.1.4.11 convert this intermediate to D-myo-inositol 1-phosphate or IP3 in a slow, phosphodiesterase-catalyzed reaction step. Phosphoinositide-specific PLCs (class EC 4.6.1.11) play an important role in modulation of phosphatidyl inositol(4,5)-bisphosphate and (3,4,5)-inositol-trisphosphate metabolism. Some of the microbial PLCs possess a C2 domain (Fig. 6) like most mammalian PLCs, which is involved in Ca2+-ion mediated binding of the enzyme to the membrane.
4. Microbial PLDs
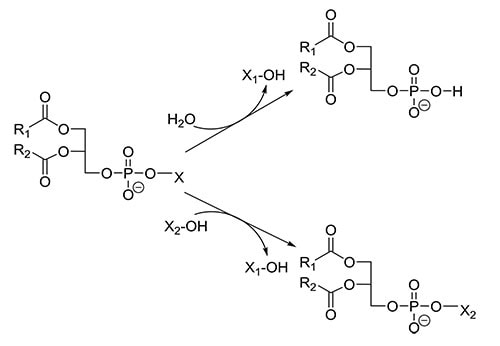
PLDs (EC 3.1.4.4) have been identified not only in the plant and animal kingdom, but also in several microorganisms. PLDs release the phospholipid head group from glycerophospholipids to yield phosphatidic acid (PA), which is involved in a number of cellular signaling processes. Due to its smaller head group, a changed membrane curvature is achieved, which is a driving force in membrane trafficking and fusion events. The downstream products of PLD, diacylglycerol (DAG) and lysophosphatidic acid (LPA) are also important lipid signaling molecules. Besides hydrolysis, PLDs catalyze the exchange of the alcohol moiety in the head group by an extraneously added alcohol in a so-called transphosphatidylation reaction (Fig. 7). Microbial PLDs are involved in the invasion of host cells; however, much remains to be studied with respect to their roles as virulence factors in pathogenesis and the life cycle of the microorganisms itself.
Figure 7: Reactions catalyzed by PLD.
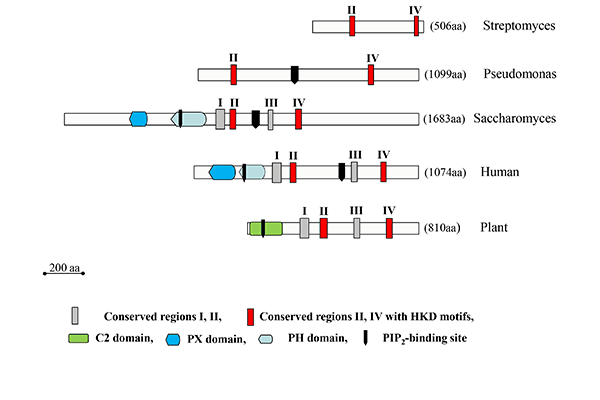
Highly homologous PLDs are secreted by several Gram-positive Streptomyces strains. Some of them actually represent the best-characterized PLDs to date and the sole microbial PLDs commercially used for the production of natural and synthetic phospholipids. This is favored by their high transphosphatidylation activities, broad substrate specificity and the availability of genetically engineered variants. Due to the lack of any regulatory domain, Streptomyces PLDs are the smallest PLDs known (Fig. 8). Unlike PLDs from plants, they do not require metal ions for enzyme activity.
The largest (in terms of size?) (yes) microbial PLDs are found in yeasts. They resemble mammalian PLDs (Fig. 8). Thus, Spo14 or PLD1 from S. cerevisiae, with a molecular mass of 195 kDa, contains, besides the N-terminal regulatory ‘localization and phosphorylation’ domain (Loco/Phosp), PX and PH domains, a polybasic PI(4,5)P2-binding domain between the two essential catalytic motifs. Spo14-like PLDs are also found in Candida albicans and Schizosaccharomyces pombe. These enzymes play an essential role in vesicle fusion events.
Figure 8: Schematic representation of the structure of microbial PLD enzymes in comparison to their plant and mammalian counterparts. The number of amino acid residues for each enzyme is indicated at the right. The human enzymes are represented by PLD1 (Q13393), and the plant enzymes by PLDα1 from Arabidopsis thaliana (AEE75720). The microbial PLDs are in the order of appearance the enzymes from S. sp. PMF (P84147), P. aeruginosa (NP_252177), and S. cerevisiae (CAA82103).
PLDs are also found in as well as Chlamydiae (pzPLD) and various other Gram-negative pathogenic bacteria, such as Neisseria gonorrhoeae (NgPLD) and Yersinia pestis, producing the well characterized toxin(Ymt, which yielded the first proof for the existence of the phospho-histidine intermediate), P. aeruginosa (PLDA, PLDB, which are highly homologous to mouse PLD2), Acinetobacter baumanii, Klebsiella, Rickettsia and L. pneumophila (LpdA) and different fungi, such as A. nidulans and A. fumigatus (PLDA), protozoa (Tetrahymena) or the slime mold Dictyostelium discoidum (PLDB). At the molecular level, most of these enzymes are only poorly characterized compared with the enzymes from Streptomyces and Yersinia strains.
Viruses contain several proteins with one HKD motif; however, activities are mostly connected with endonuclease (Nuc) or highly unspecific phospholipid (p37 lipase) cleavage activity.
PLDs belong to a large superfamily of proteins all of which contain one or two of the highly conserved HxK(x)4D(x)6G(S/G) motifs first described by Ponting and Kerr, and share a common catalytic mechanism even though their substrates are not in all cases phospholipids. The HKD motifs, which are located in the case of PLDs in the conserved regions II and IV (Fig. 8), are essential for catalysis and two of them comprise the active site, as clearly shown by several crystal structures of enzymes belonging to this superfamily.
For two PLDs secreted by the Gram-positive Streptomyces strains, Streptomyces spec. PMF and S. antibioticus, crystal structures are available (PDB codes 1F01 and 2ZE4, respectively). Details of the previously suggested ping-pong mechanism, resulting in the formation of a phosphatidyl-enzyme intermediate, could be derived by resolution of the crystal structures in the presence of short-chain phospholipids and tungstate. The phosphatidyl-enzyme intermediate is formed by nucleophilic attack of the His residue in the N-terminal HKD motif at the phosphodiester bond. The His residue of the C-terminal HKD motif functions as general acid and protonates the leaving group, the corresponding alcohol.
PLDs not belonging to this superfamily are PLDs from Corynebacterium pseudotuberculosis, Arcanobacterium haemolyticus, Shewanella spp., and especially from Streptomyces chromofuscus. The last one is still erroneously considered in research as model enzyme for PLD when studying membrane binding, interfacial activation, and transphosphatidylation even though its sequence characteristics, properties, reaction mechanism, and metal ion requirement are highly divergent to those of the HKD-type enzymes. These enzymes closely resemble purple acid phosphatases and require Fe3+ and Mn2+ for activity and might be grouped into the PhoD (phosphodiesterase/alkaline phosphatase) family.
Genes encoding N-acylphosphatidylethanolamine (NAPE)-specific PLD were predicted for S. cerevisiae and Colletotrichum. Significant amounts of N–acylethanolamines (NAEs) and NAPEs were also found in distinct microorganisms.
Summary
Microbial phospholipases exhibit a great diversity with respect to sequence characteristics, biochemical properties, mode of action, biological function within the microorganism itself, and their impact on pathogenesis of the individual microorganism. Knowledge on mechanistic details and the role in pathological processes of many of the phospholipases is still rather limited.
Microbial phospholipases act as virulence factors triggering internalization into host cells and beyond that also the destruction of bacterial competitors. Further studies in this field might shed light on the role that each individual phospholipase plays within the microorganism itself, as well as in its immediate environment. Improved knowledge in this field might be helpful for the rational development of specific inhibitors to overcome the burdens of pathogenic microorganisms on humans. Genome sequencing and sequence comparisons might be valuable tools to obtain a more general picture of these microbial enzymes and help our understanding of the functions of the individual phospholipases.
The author wishes to thank Prof. Renate Ulbrich-Hofmann, Institute of Biochemistry and Biotechnology for critical reading of the manuscript and discussions, and Prof. Gary Sawers, Institute of Biology, Martin-Luther University Halle for reading the manuscript.
Recommended reading:
- Ghannoum, M. Potential Role of Phospholipases in Virulence and Fungal Pathogenesis. Clin. Microbiol. Rev., 13, 122-143 (2000).
- Schaloske, R.H. and Dennis, E.A. The phospholipase A2 superfamily and its group numbering system. Biochim. Biophys. Acta, 1761,1246-59 (2006).
- Köhler, G.A., Brenot, A., Haas-Stapleton, E., Agabian, N., Deva, R. and Nigam, S. Phospholipase A2 and phospholipase B activities in fungi. Biochim. Biophys. Acta, 1761, 1391-9 (2006).
- Dekker, N. Outer-membrane phospholipase: known structure, unknown biological function. Mol. Microbiol., 35, 711-7 (2000).
- Matoba, A. and Sugiyama, M. Atomic resolution structure of prokaryotic phospholipase A2: analysis of internal motion and implication for a catalytic mechanism. Proteins, 15, 453-69 (2003).
- Anderson, D.M., Sato, H., Dirck, A.T., Feix, J.B. and Frank, D.W. Ubiquitin activates patatin-like phospholipases from multiple bacterial species. J. Bacteriol., 197, 529-41 (2015).
- Hong, S.H., Horiuchi, H. and Ohta, A. Identification and molecular cloning of a gene encoding Phospholipase A2 (plaA) from Aspergillus nidulans. Biochim. Biophys. Acta, 1735, 222-9 (2005).
- Russell, A.B., Leroux, M., Hathazi, K., Agnello, D.M. Ishikawa, T., Wiggins, P.A., Wai, S.N. and Mogous, J.D. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature, 496, 508-12 (2013).
- Djordjevic, J.T. Role of phospholipases in fungal fitness, pathogenicity, and drug development – lessons from Cryptococcus neoformans. Front. Microbiol. (2010) (doi: 10.3389/fmicb.2010.00125).
- Titball, R.W. Bacterial phospholipases C. Microbiol. Rev., 57, 347-66 (1993).
- Griffith, O.H. and Ryan, M. Bacterial phosphatidylinositol-specific phospholipase C: structure, function, and interaction with lipids. Biochim. Biophys. Acta, 1441, 237-54 (1999).
- Heinz, D.W., Essen, L.O. and Williams, R.L. Structural and mechanistic comparison of prokaryotic and eukaryotic phosphoinositide-specific phospholipases C. J. Mol. Biol., 275, 635–50 (1998).
- Katan, M. Families of phosphoinositide-specific phospholipase C: structure and function. Biochim. Biophys. Acta, 1436, 5-17 (1998)
- Ponting, C.P. and Kerr, I.D. A novel family of phospholipase D homologues that includes phospholipid synthases and putative endonucleases: identification of duplicated repeats and potential active site residues. Protein Sci., 5, 914-22 (1996).
- Selvy, P.E., Lavieri, R.R., Lindsley, C.W. and Brown, H.A. Phospholipase D: enzymology, functionality, and chemical modulation. Chem. Rev., 111, 6064-119 (2011).
- Mansfeld, J. and Ulbrich-Hofmann, R. Modulation of phospholipase D activity in vitro. Biochim. Biophys. Acta, 1791, 913-26 (2009).
- Uesugi, Y. and Hatanaka, T. Phospholipase D mechanism using Streptomyces PLD. Biochim. Biophys. Acta, 1791, 962-9 (2009).
- Mendonsa, R. and Engebrecht, J. Phospholipase D function in Saccharomyces cerevisiae. Biochim. Biophys. Acta, 1791, 970-4 2009.
Related Resources
Lipid Library
Edible Oil Processing
In the present context, the term edible oil processing covers the range of industrial…
Lipid Library
The Highs and Lows of Cannabis Testing
October 2016 With increasing legalization of both adult recreational and medical cannabis,…
Lipid Library
The secrets of Belgian chocolate
By Laura Cassiday May 2012 Like a bonbon nestled snugly in a…