1. Commercial utilization of bacterial lipids
Lipids are valuable compounds for versatile commercial applications and have the potential to replace fossil resources in many industrial processes. There has been made remarkable progress regarding the utilization of oils as sustainable alternative feedstock to generate a multitude of basic oleochemicals for organic synthesis, catalysis or biotechnology in the chemical industry [1]. A major global objective, however, is the replacement of fossil fuels in the transportation sector and, to date, biodiesel made from plant, animal or microbial oils is the predominant biofuel [2].
Conventional biodiesel is produced by transesterification of triacylglycerols (TAG) from vegetable oil with short chain alcohols – usually methanol or ethanol. However, the oil yield of common oil crops is very poor, for example, canola yields only 1200 L/ha and oil palm about 6000 L/ha because the plant’s oil contents are often less than 5% of their biomass. Another drawback is the immense need for arable land, which inevitably conflicts with the world food supply. Some microorganisms reach much higher oil contents (of up to roughly 70% of their cell dry weight; CDW) but do not require fertile land and only a fractional amount of space for their cultivation. It is thus inevitable to establish alternative oil sources to replace fossil fuels in a more sustainable, and – in the long term –economically feasible way. This could especially be achieved, if the microorganisms utilized cheap industrial by-products, waste compounds, or plant material which can be produced on rather barren land (so-called “energy crops”). Microorganisms are thus seen as promising sources for high-value lipids and provide significant advantages over traditionally used oils of plant or animal origin. Bacteria have a less complex genome, metabolism, and cell compartmentation and can produce a wider range of different types of lipids compared to multicellular eukaryotes. In addition, they can usually be genetically modified more readily to obtain optimized strains that further increase the productivity and competitiveness of the whole process [2–4].
Wax esters (WE), composed of long chain alcohol and fatty acid residues, represent another type of lipid which is usually not accumulated by eukaryotes in significant amounts. WE are valuable ingredients of medical or cosmetic products, lubricants or coatings, amongst others. Usually, they are harvested from plants, such as jojoba or carnauba wax, which require rather time-consuming and expensive cultivation. Certain bacteria can offer an interesting alternative source of WE with a similar composition compared to jojoba oil. Furthermore, the desired composition can be targeted by the choice of precursor substrates [5, 6].
According to the current state of knowledge, noteworthy bacterial TAG or WE synthesis is catalyzed by the wax ester synthase/acyl-CoA:diacylglycerol acyltransferase (WS/DGAT). Although alternative routes for TAG synthesis exist in some bacterial species, they usually do not account for a significant accumulation of this storage compound [7].
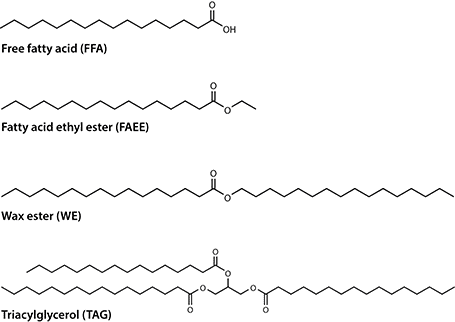
Other bacterial lipids, such as polyesters (polyhydroxyalkanoates, PHA) or rhamnolipids, also have a great industrial relevance as bioplastic or biosurfactants, respectively. However, these areas are beyond the scope of this article, which focuses on bacteria producing neutral and non-polymeric lipids such as FFA, fatty acid ethyl esters (FAEE), WE or TAG (Fig. 1).
Fig. 1 Exemplary chemical structures of industrially relevant lipids produced by bacteria.
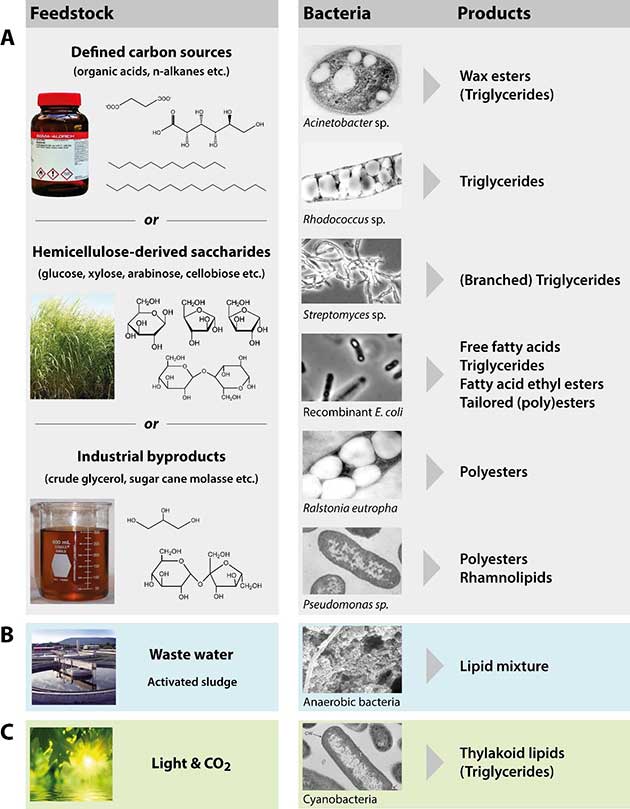
Figure 2 gives a schematically overview of different strategies to employ bacteria to convert different types of feedstock into valuable lipids for industrial or commercial applications. While defined carbon sources might only be considered for very high-value end products (such as for medical applications), much cheaper substrates, like (hemi)cellulose-derived sugars or industrial waste- or by-products are necessary to decrease production costs for bulk lipids (as required for biodiesel production) and to increase the overall sustainability of the process (Fig. 2 A). Other very promising approaches aim at converting waste water by the bacterial community in activated sludge (Fig. 2 B) and CO2 by microalgae or cyanobacteria (Fig. 2 C) to generate lipids. The following sections will discuss possible feedstock as well as promising bacterial production strains, either naturally capable of, or genetically engineered to accumulate lipids.
Fig. 2 Strategies to employ bacteria for the production of industrially relevant lipids from diverse feedstock.
2. Feedstock for the production of bacterial lipids
The utilization of sucrose- or starch-derived sugars for a large-scale bacterial production of lipids is economically and ethically problematic. Many types of lipids are usually produced most efficiently from structurally related substrates like alkanes and alkanoic acids. However, their low miscibility and toxicity, respectively, and their high prices make an economic production with these substrates problematic and unlikely. As the substrate cost can contribute up to 50% of the total production cost, it greatly affects the price competitiveness of lipid-derived fuels compared to conventional fuel. Hence, the most obvious step in order to minimize the production cost is to employ cheap carbon and/or nitrogen sources from agricultural or industrial waste and surplus materials, such as hydrolyzed plant biomass, sugar-rich molasses, glycerol from biodiesel production or whey as complex nitrogen and phosphorus source [4, 8, 9].
Lignocellulose from agricultural, industrial and forest residuals represents the worldwide largest and cheapest resource of sugars with about 4.15 billion tons of various types of agricultural waste accruing annually. Besides, the utilization of this low cost and abundant feedstock would not only make lipid production ecologically sound, but also geopolitically independent from oil-producing countries. However, due to the complex and stable structure of lignocellulose, its breakdown into fermentable sugars requires a time- and energy-consuming pretreatment. Therefore, it would be advantageous to employ bacterial strains that either degrade (hemi)cellulose or efficiently use the released hexoses and pentoses, such as D-xylose, L-arabinose, D-glucose, D-mannose, and D-galactose to produce so-called “second-generation biofuels” [10–12].
Other interesting approaches, which are currently discussed, aim at establishing effective processes to gain microbial lipids produced from organic waste or carbon dioxide for the production of “third-generation biofuels”. During wastewater treatment, specialized anaerobic microorganisms accumulate lipid mixtures composed of TAG, WE or PHA. It is estimated that extraction and transesterification of these lipids in 50% of all biological wastewater treatment plants of the U.S. could cover 0.5% of its yearly diesel demand, and thus could play a role in a future energy strategy which utilizes as many renewable resources as possible [13]. Oleaginous photosynthetic microorganisms can efficiently convert solar energy and recycle carbon dioxide into lipids. While microalgae store TAG under environmental stress, some cyanobacteria accumulate significant amounts of lipids in the form of diacylglycerol in their thylakoid membranes and could even be genetically engineered to overproduce FFA [14].
3. Bacterial lipid producers
When evaluating bacterial strains regarding a potential utilization as industrial lipid producer, several important factors should be taken into account: i, expensive carbon sources and media supplements should not be necessary; ii, the growth rates, achievable cell densities and product concentrations, especially under fermentation conditions, should be high; iii, while the extraction of the product should be as easy as possible. An extracellular product would enable a continuous cultivation and reduce extraction costs. Furthermore, the bacteria must be non-pathogenic and genetically accessible if directed strain optimization is necessary.
3.1. Lipid accumulation in Gram-positive bacteria
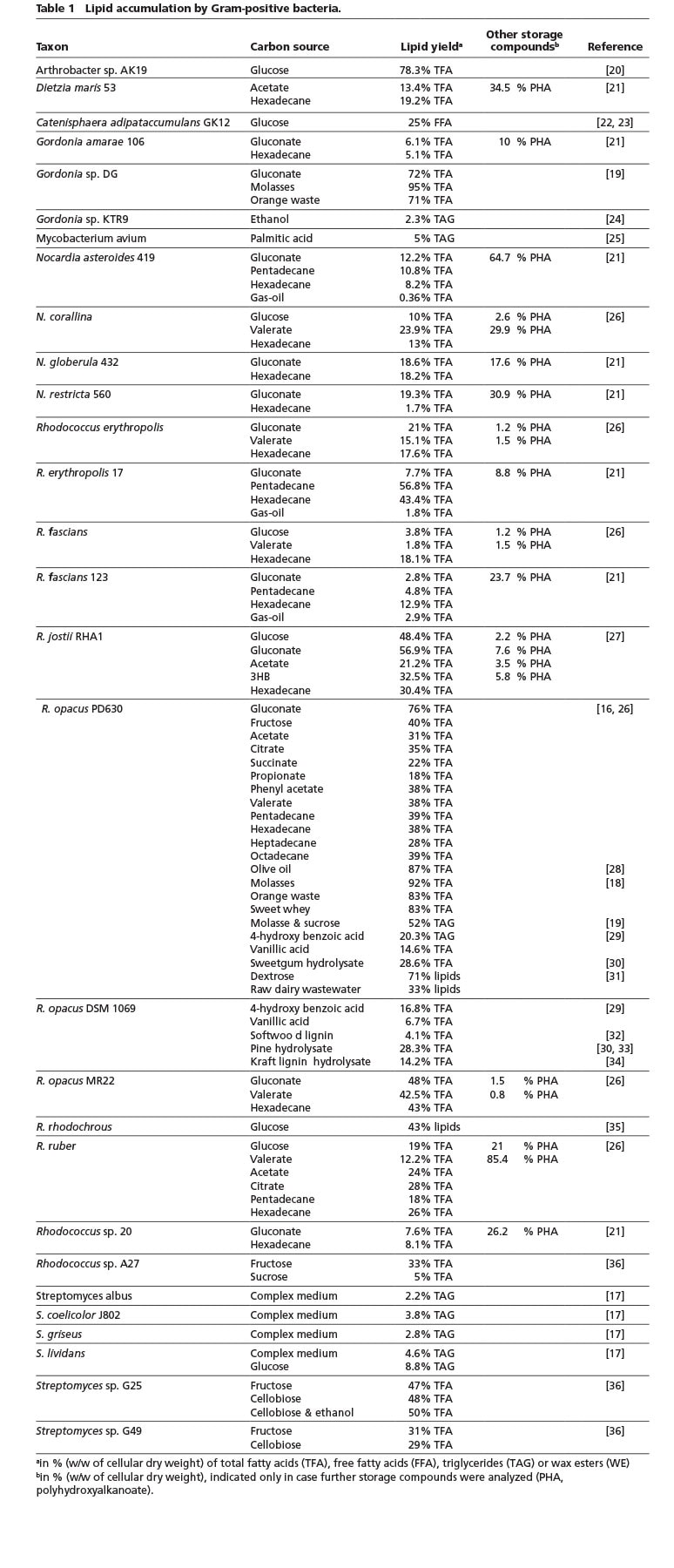
The ability to accumulate significant amounts of TAG as intracellular storage deposits is predominantly found in bacteria belonging to the Gram-positive Actinomycetales, e. g. Arthrobacter, Dietzia, Gordonia, Nocardia, Rhodococcus or Streptomyces species (Table 1). Thereof, especially R. opacus PD630 is widely known for its impressive TAG contents of up to around 80% of its CDW and seen as a very promising strain for industrial TAG production due to its fast growth, acceptance of a wide range of carbon sources and high substrate tolerance [15]. Although Arthrobacter AK19 was reported many years ago to accumulate similar high amounts, this strain has not been used in further studies. Rhodococcus or Gordonia strains especially appear suitable for the conversion of industrial waste materials (such as molasses, wastes from different agro-industrial processes, dairy wastewater etc.) or lignocellulose hydrolysates, while a Streptomyces isolate can utilize cellobiose to build up to 50% fatty acids of its CDW (Table 1). In contrast to TAG synthesized by Rhodococcus, which are mainly composed of straight-chain C16 – C18 fatty acid residues, streptomycetes have the special feature to also incorporate branched-chain fatty acids [16, 17].
It is important to note that lipid contents are often only measured in batch cultivation, although continuous cultivation could significantly increase the overall lipid productivity. However, a high cellular content of lipids would not automatically make the process economically feasible, as the overall lipid yield might still be comparably low (e.g. although the lipid content after batch cultivation of R. opacus cells with molasses was reported to be 92% of its CDW, only around 55 mg/L, corresponding to a productivity of approx. 0.6 mg/L·h were reached [18]). On the contrary, in a 30 L fed-batch fermentation the productivity of R. opacus was significantly increased to 19.4 g/L or 0.38 g/L·h, respectively, of total fatty acids (corresponding to approx. 50% of its CDW) from molasses and sucrose [19]. Thus, individual cultivations and the achieved lipid yields are not necessarily comparable.
The Firmicutes isolate Catenisphaera adipataccumulans GK12 was reported to accumulate FFA intracellularly in amounts of up to 25% of CDW – a phenomenon which has not been observed in nature, so far [22]. However, it is questionable, if the lipids were accumulated in the form of FFA, which are very cytotoxic, even at relatively low concentrations. Alternatively, FFA could have been released from accumulated lipids during the processing of the cells. Furthermore, Streptomyces sp. NP10 showed the rather unusual characteristic of secreting large amounts of FFA with a complex composition when grown on complex medium [37].
In activated sludge of biological wastewater treatment plants there typically can be found a high proportion of actinomycetes in the foam of the anoxic tank. This complex consortium of oleaginous bacteria could be simply skimmed from the surface, which would facilitate the recovery of this lipid-rich biomass for biodiesel production [13].
3.2. Lipid accumulation in Gram-negative bacteria
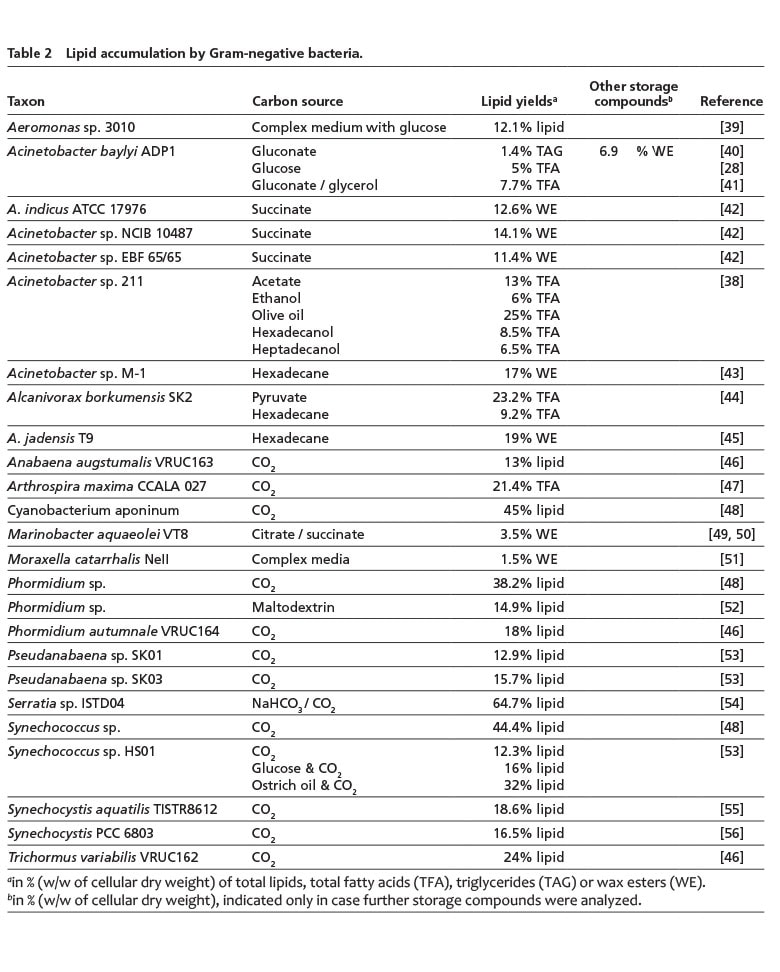
In Gram-negative bacteria, TAG storage in comparable amounts as achieved by some Gram-positive bacteria has not yet been reported (Table 2). Instead, some Gram-negative bacteria, in particular Acinetobacter or hydrocarbonoclastic bacteria, such as Alcanivorax or Marinobacter species, are able to synthesize WE, which are then often accumulated with lower amounts of TAG in mixed lipid inclusions. Noteworthy amounts are usually only achieved, when these bacteria are cultivated with related carbon sources, like n-alkanes or olive oil [6, 38].
An interesting Gram-negative isolate, Aeromonas sp. 3010, achieved a total lipid content of approx. 12%, whereof up to 30% is eicosapentaenoic acid [20:5 n-3, EPA], a polyunsaturated fatty acid with great relevance for pharmaceutical or food industries (Table 2). Furthermore, two rather unusual findings of lipid-producing Gram-negative bacteria have been reported: An isolate assigned to the genus Nitratireductor (an α-proteobacterium), which could be interesting for waste-water treatment, grows with short-chain organic acids and synthesizes an unusual mixture of lipids composed of squalene, TAG and light oils (such as 2-butenoic acid methyl ester) with contents of up to 70% of the CDW [57]. In addition, a chemolithotrophic, CO2-sequestering Serratia strain synthesized approx. 50% of its CDW as long-chain hydrocarbons and lipids, and up to 466 mg extracellular lipids/L, respectively [54, 58]. However, there are also reports on oleaginous Pseudomonas and Bacillus strains [59, 60] that require further validation as it is rather unlikely that these species are capable of accumulating neutral lipids but are instead known for their ability to store PHA. These isolates might not have been correctly assigned at the taxonomic level. Furthermore, the occurrence of neutral lipids in P. aeruginosa 44T1 was not be confirmed in an independent study [61].
Cyanobacteria may represent another special case, as storage of intracellular TAG has not yet been established in this group of microorganisms. There is no (WS/)DGAT gene present in published cyanobacterial genomes. However, their photosynthetic membranes can serve as large reservoirs for membrane lipids and diacylglycerols, which could also be utilized biotechnologically, e.g. for transesterification with methanol to yield biodiesel [14, 53]). Using CO2 as sole carbon source, some cyanobacterial strains have reached impressive lipid contents of more than 40% of the CDW, such as Cyanobacterium aponinum or Synechococcus sp. (Table 2) but these lipids are complex entities that would need hydrolysis to release the required fatty acids.
For a bulk production of lipids, e.g. as feedstock for biodiesel, especially bacteria belonging to the actinomycetes, such as certain Rhodococcus or Streptomyces species, represent robust production strains that are capable of growing on cheap raw materials and can accumulate high amounts of TAG. Although WE-synthesizing Gram-negative bacteria usually do not achieve similar high lipid contents and require more expensive carbon sources, some WEs with desirable compositions can be produced for rather high-value end products, such as cosmetics.
Although promising production strains already could and will be isolated from natural habitats, genetic engineering has a great potential to further optimize these strains regarding their lipid accumulation and also their substrate spectrum. In addition, non-oleaginous bacteria, like E. coli, can be transformed into valuable production strains by means of genetic engineering to overproduce FFA, TAG or FAEE, as outlined below.
3.3. Free fatty acid overproduction in genetically-engineered bacteria
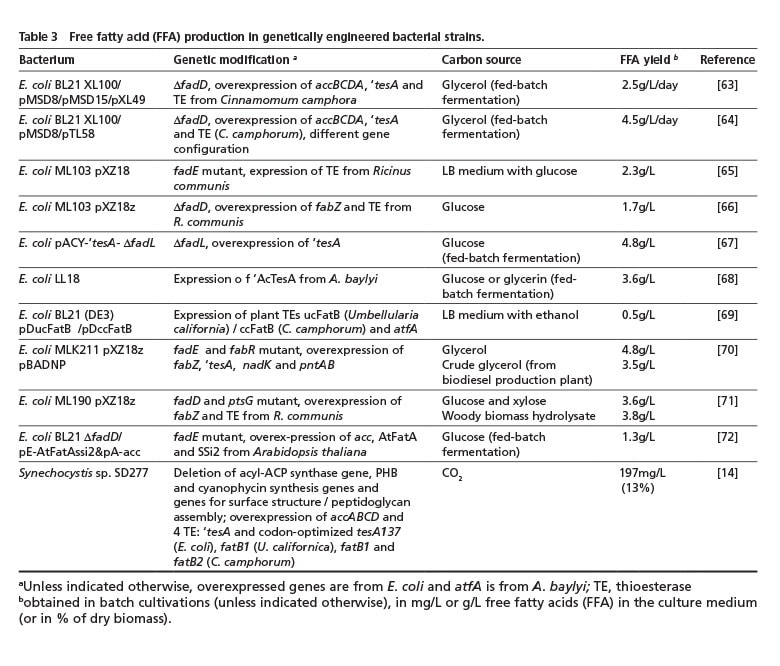
Except for the aforementioned rare findings of an accumulation or excretion of FFA by natural isolates, FFAs are usually not overproduced and do not represent an intrinsic storage compound due to their toxicity compared to TAG or WE [62]. Remarkable process has been made during the last decade regarding genetic optimization of E. coli as well as cyanobacteria to overproduce and secrete FFAs (Table 3).
The secretion of fatty acids into the cultivation medium would eliminate the effort of harvesting, drying and solvent-extracting lipid-containing cells and, thus, greatly reduce the overall production costs of bacterial lipids. Although not naturally capable of accumulating lipids, E. coli is an attractive production organism as its lipid metabolism has been examined in the most detail amongst prokaryotes and numerous tools have been well established for its genetic modification. Thus, by overexpression of genes involved in fatty acid de novo synthesis and/or blocking genes of fatty acid degradation, significant fatty acid overproduction can be achieved. The overexpression of (native or recombinant) thioesterases then releases the acyl residue from acyl binding protein (ACP) and FFA accumulate [62]. Up to 4.8 g FFA/L has already been obtained by engineered E. coli strains using glycerol or glucose in fed-batch fermentation, or even 3.8 g/L from woody biomass hydrolysate (Table 3).
The cyanobacterium, Synechocystis sp. SD277, was extensively modified by deleting several genes involved in competing processes (such as the undesirable synthesis of PHB or cyanophycin instead of fatty acids) or to weaken the cell wall integrity to facilitate FFA release into the medium. After overexpressing several thioesterase genes, this engineered strain produced up to 200 mg FFA/L [14] (Table 3). Although the yield is far below the values obtained with E. coli, this process would nevertheless be much more sustainable because CO2 can serve as the sole carbon source.
3.4. Triacylglycerol synthesis in genetically-engineered bacteria
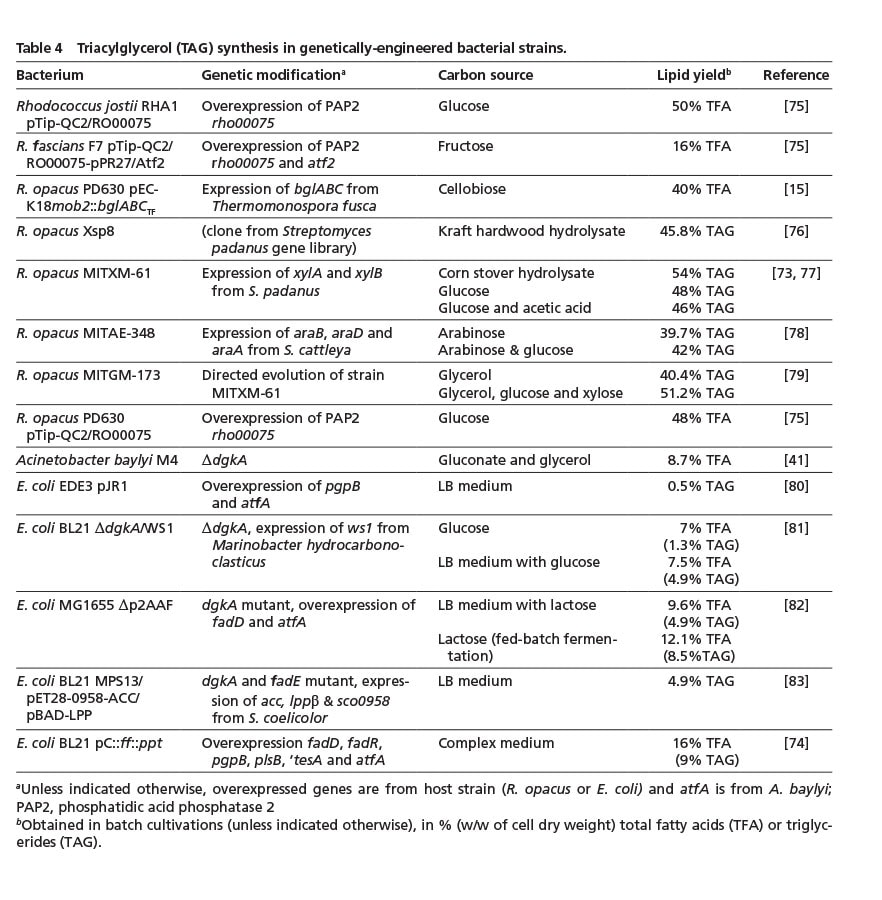
As mentioned earlier, R. opacus can accumulate large amounts of TAG. Thus, most genetic engineering approaches have been aimed at extending its substrate range to enable a more sustainable lipid production from glycerol, (hemi)cellulose-derived sugars or hydrolyzed plant material (Table 4). By means of targeted modifications and an adaptive evolution strategy, more than 50% (w/w) TAG (corresponding to ca. 16 g/L) was achieved using corn stover hydrolysate as sole carbon source [73]. These yields are significantly above the amounts obtained by wild-type R. opacus with different hydrolyzed plant materials (of approx. 28% fatty acids; Table 1) which clearly underlines the importance of strain optimization for a competitive bacterial lipid production from a challenging feedstock.
A number of studies also focused on establishing TAG accumulation in E. coli; for example by combining increased fatty acid and diacylglycerol synthesis with a recombinant expression of essential WS/DGAT genes [74] (Table 4). However, the achievable yields still need to be significantly improved in order to compete with natural lipid accumulators (Table 1 & 4).
3.5. Fatty acid ethyl ester synthesis in genetically-engineered bacteria
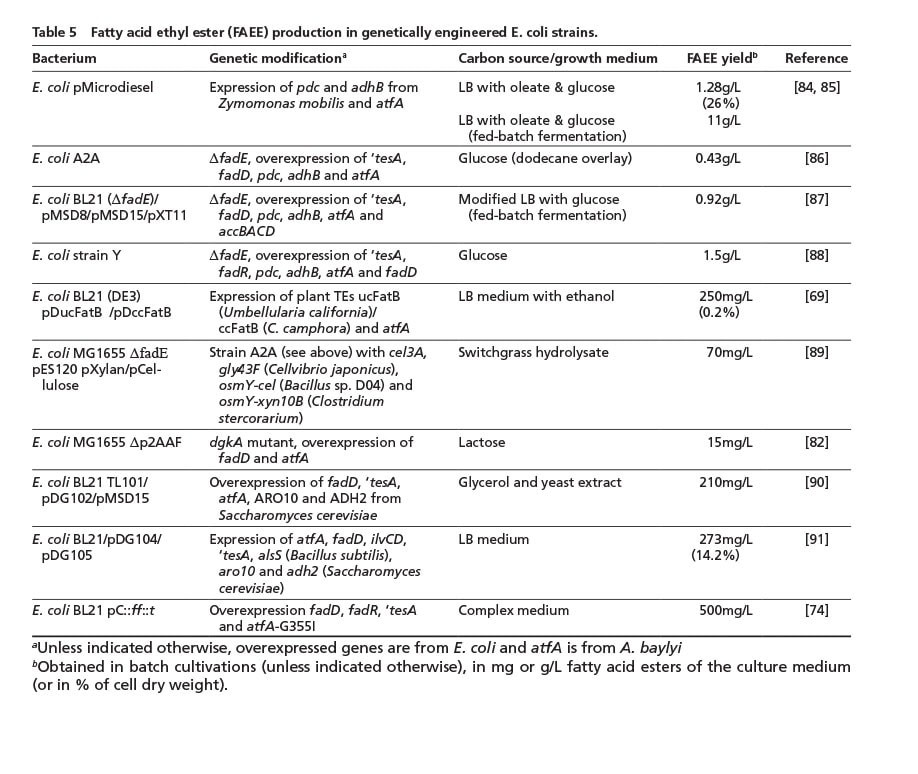
The vast majority of biodiesel (fatty acid methyl ester, FAME) is currently produced with methanol derived from fossil sources, although ethanol can alternatively be utilized to gain biodiesel made from fatty acid ethyl esters (FAEE) which can show superior physicochemical properties compared to FAME [8]. In 2006, a strategy was developed to completely synthesize FAEE de novo (“microdiesel”) from unrelated carbon sources in E. coli [84]. In this first approach and follow-up studies, the overexpression of a WS/DGAT gene was usually combined with the ethanol synthesis pathway from Zymomonas mobilis (adhB encoding alcohol dehydrogenase B, and pdc for pyruvate decarboxylase). Similar to strategies employed for FFA overproduction, increasing fatty acid de novo synthesis and blocking the beta-oxidation of fatty acids, could further increase FAEE levels in more recent studies (Table 5).
So far, highest FAEE amounts of 1.5 g/L were only achieved with glucose and in small-scale cultivations. A major disadvantage of E. coli is its inability to metabolize lignocellulose, although a first proof-of-principle could be achieved by constructing an optimized strain which utilizes switchgrass hydrolyzate [86] (Table 5).
Another promising approach is the synthesis of branched-chain fatty acid alkyl esters from (branched) fatty acids and branched short-chain alcohols (e.g. isoamyl alcohol or isobutanol), exploiting the biosynthesis pathway for branched-chain amino acid synthesis. These esters show improved properties at low temperatures, under which conventional biodiesel usually has a rather poor performance [91] (Table 5). However, the synthesis yield of branched FAEE still needs to be considerably increased. Alternatively, bacteria which naturally incorporate branched fatty acids into TAGs, such as Streptomyces isolate G25 (Table 1), could be advantageous for the production of biodiesel with a mixture of straight- and branched-chain fatty acid residues.
To sum up, an economically feasible production of bacterial lipids ideally requires robust and fast-growing strains. These bacteria should not only achieve high lipid yields but should also be able to utilize abundant and low-cost feedstocks, such as industrial waste- or surplus materials. During the last decade, there has been made considerable progress regarding the genetic engineering of bacteria to increase their lipid contents or substrate ranges. However, these modified strains could not yield lipid yields comparable to those of some outstanding oleaginous wild-type bacteria, such as R. opacus, yet. Metabolic engineering appears to be a promising strategy to enable the synthesis of rather artificial lipids, such as FAEE or tailor-made, wax ester-like compounds for more specific purposes. Furthermore, it could promote a more complete utilization of the whole spectrum of carbon sources which are present in – for example – lignocellulosic hydrolysates or might increase the robustness of a production strain towards certain inhibitors. An impressive amount of proof-of-principle studies have already been conducted, so far, which can lay the foundation for targeted modifications to further improve the most promising production strains. To date, these are R. opacus or Streptomyces sp. G25 for bulk production of TAG from lignocellulose-derived saccharides, certain cyanobacteria for a CO2-based production of lipids, as well as recombinant E. coli strains for the synthesis of FAEE-biodiesel.
Acknowledgment
The authors are very grateful to the Rahn-Quade-Stiftung for financial support by a fellowship to Annika Röttig (Project T381 of the Deutsches Stiftungszentrum in Essen).
4. References
- Biermann U, Bornscheuer U, Meier MAR, Metzger JO, Schäfer HJ (2011) Oils and fats as renewable raw materials in chemistry. Angew Chem Int Ed 50:3854–3871
- Kosa M, Ragauskas AJ (2011) Lipids from heterotrophic microbes: advances in metabolism research. Trends Biotechnol 29:53–60.
- Chisti Y (2007) Biodiesel from microalgae. Biotechnol Adv 25:294–306
- Rude MA, Schirmer A (2009) New microbial fuels: a biotech perspective. Curr Opin Microbiol 12:274–281
- Alvarez HM (2010) Biotechnological production and significance of triacylglycerols and wax esters, in: Timmis, K.N. (Ed.), Handbook of Hydrocarbon and Lipid Microbiology. Springer Berlin Heidelberg, 2995–3002.
- Ishige T, Tani A, Sakai Y, Kato N (2003) Wax ester production by bacteria. Curr Opin Microbiol 6:244–250.
- Röttig A, Steinbüchel A (2013) Acyltransferases in bacteria. Microbiol Mol Biol Rev 77:277–321.
- Röttig A, Wenning L, Bröker D, Steinbüchel A (2010) Fatty acid alkyl esters: perspectives for production of alternative biofuels. Appl Microbiol Biotechnol 85:1713–1733.
- Sun Z, Ramsay JA, Guay M, Ramsay BA (2007) Fermentation process development for the production of medium-chain-length poly-3-hydroxyalkanoates. Appl Microbiol Biotechnol 75:475–485.
- Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD (2007) Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315:804–880.
- Kim J, Yun S (2006) Discovery of cellulose as a smart material. Macromolecules 39:4202–4206.
- Stephanopoulos G (2007) Challenges in engineering microbes for biofuels production. Science 315:801–804.
- Muller EEL, Sheik AR, Wilmes P (2014) Lipid-based biofuel production from wastewater. Curr Opin Biotechnol 30:9–16.
- Liu X, Sheng J, Curtis III R (2011) Fatty acid production in genetically modified cyanobacteria. Proc Natl Acad Sci 108:6899–6904.
- Hetzler S, Steinbüchel A (2013) Establishment of cellobiose utilization for lipid production in Rhodococcus opacus PD630. Appl Environ Microbiol 79:3122–3125.
- Alvarez HM, Mayer F, Fabritius D, Steinbüchel A (1996) Formation of intracytoplasmic lipid inclusions by Rhodococcus opacus strain PD630. Arch Microbiol 165:377–386.
- Olukoshi ER, Packter NM. 1994. Importance of stored triacylglycerols in Streptomyces: Possible carbon source for antibiotics. Microbiology (SGM)140:931–943.
- Gouda MK, Omar SH, Aouad LM (2008) Single cell oil production by Gordonia sp. DG using agro-industrial wastes. World J Microbiol Biotechnol 24:1703–1711.
- Voss I, Steinbüchel A (2001) High cell density cultivation of Rhodococcus opacus for lipid production at a pilot-plant scale. Appl Microbiol Biotechnol 55:547–555.
- Wayman M, Jenkins AD, Kormendy AG (1984) Bacterial production of fats and oils. In: Ratledge C, Dawson P, Rattray J (eds) Biotechnology for the Oils and Fats Industry. American Oil Chemists’ Society (AOCS).
- Alvarez HM (2003) Relationship between β-oxidation pathway and the hydrocarbon-degrading profile in Actinomycetes bacteria. Int Biodeterior Biodegrad 52:35–42.
- Katayama T, Kanno M, Morita N, Hori T, Narihiro T, Mitani Y, Kamagata Y (2014) An oleaginous bacterium that intrinsically accumulates long-chain free fatty acids in its cytoplasm. Appl Environ Microbiol 80:1126–1131
- Kanno M, Katayama T, Morita N, Tamaki H, Hanada S, Kamagata Y (2015) Catenisphaera adipataccumulans gen. nov., sp. nov., a member of the family Erysipelotrichaceae isolated from an anaerobic digester. Int J Syst Evol Microbiol 65:805–810.
- Eberly JO, Ringelberg DB, Indest KJ (2013) Physiological characterization of lipid accumulation and in vivo ester formation in Gordonia sp. KTR9. J Ind Microbiol Biotchnol 40:201–208.
- McCarthy C (1971) Utilization of palmitic acid by Mycobacterium avium. Infect Immun 4:199–204.
- Alvarez HM, Kalscheuer R, Steinbüchel A (1997a) Accumulation of storage lipids in species of Rhodococcus and Nocardia and effect of inhibitors and polyethylene glycol. Fett/Lipid 99:239–246.
- Hernández MA, Mohn WW, Martínez E, Rost E, Alvarez AF, Alvarez HM (2008) Biosynthesis of storage compounds by Rhodococcus jostii RHA1 and global identification of genes involved in their metabolism. BMC Genomics 9:600–614.
- Wältermann M, Hinz A, Robenek H, Troyer D, Reichelt R, Malkus U, Galla HJ, Kalscheuer R, Stöveken T, von Landenberg P, Steinbüchel A (2005) Mechanism of lipid-body formation in prokaryotes: how bacteria fatten up. Mol Microbiol 55:750–763.
- Kosa M, Ragauskas AJ (2012) Bioconversion of lignin model compounds with oleaginous Rhodococci. Appl Microbiol Biotechnol 93:891–900.
- Wei Z, Zeng G, Huang F, Kosa M, Sun Q, Meng X, Huang D, Ragauskas AJ (2015a) Microbial lipid production by oleaginous Rhodococci cultured in lignocellulosic autohydrolysates. Appl Microbiol Biotechnol 99:7369–7377.
- Kumar S, Gupta N, Pakshirajan K (2015) Simultaneous lipid production and dairy wastewater treatment using Rhodococcus opacus in a batch bioreactor for potential biodiesel application. J Environ Chem Engin 3:1630–1636.
- Kosa M, Ragauskas AJ (2013) Lignin to lipid bioconversion by oleaginous Rhodococci. Green Chem 15:2070–2074.
- Wei Z, Zeng G, Huang F, Kosa M, Huang D, Ragauskas AJ (2015b) Bioconversion of oxygen-pretreated Kraft lignin to microbial lipid with oleaginous Rhodococcus opacus DSM 1069. Green Chem 17:2784–2789.
- Wells T Jr., Wei Z, Ragauskas (2015) Bioconversion of lignocellulosic pretreatment effluent via oleaginous Rhodococcus opacus DSM 1069. Biomass Bioenerg 72:200–205.
- Shields-Menard SA, Amirsadeghi M, Sukhbaatar B, Revellame E, Hernandez R, Donaldson JR, French WT (2015) Lipid accumulation by Rhodococcus rhodochrous grown on glucose: J Ind Microbiol Biotechnol 42:693–699.
- Röttig A, Hauschild P, Madkour MH, Al-Ansari AM, Almakishah NH, Steinbüchel A (2016) Isolation of lipid-accumulating bacteria from desert soil. Submitted for publication
- Ilic-Tomic T, Genčić, Živković, Vasiljevic B, Djokic L, Nikodinovic-Runic J, Radulović (2015) Structural diversity and possible functional roles of free fatty acids of the novel soil isolate Streptomyces sp. NP10. Appl Microbiol Biotechnol 99:4815–4833.
- Alvarez HM, Pucci OH, Steinbüchel A (1997b) Lipid storage compounds in marine bacteria. Appl Microbiol Biotechnol 47:132–139.
- Woong Cho K, Jun Mo S (1999) Screening and characterization of eicosapentaenoic acid-producing marine bacteria. Biotechnol Lett 21:215–218.
- Kalscheuer R, Steinbüchel A (2003) A novel bifunctional wax ester synthase/acyl-CoA:diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1. J Biol Chem 278:8075–8082.
- Santala S, Efimova E, Kivinen V, Larjo A, Aho T, Karp M, Santala V (2011) Improved triacylglycerol production in Acinetobacter baylyi ADP1 by metabolic engineering. Microbial Cell Factories 10:36–46.
- Fixter LM, Nagi MN, McCormack JG, Fewson CA (1986) Structure, distribution and function of wax esters in Acinetobacter calcoaceticus. J Gen Microbiol 132:3147–3157.
- Ishige T, Tani A, Takabe K, Kawasaki K, Sakai Y, Kato N (2002) Wax Ester Production from n-alkanes by Acinetobacter sp. strain M-1: ultrastructure of cellular inclusions and role of acyl Coenzyme A reductase. Appl Environ Microbiol 68:1192–1195.
- Kalscheuer R, Stöveken T, Malkus U, Reichelt R, Golyshin PN, Sabirova JS, Ferrer M, Timmis KN, Steinbüchel A (2007) Analysis of storage lipid accumulation in Alcanivorax borkumensis: evidence for alternative triacylglycerol biosynthesis routes in bacteria. J Bacteriol 189:918–928.
- Bredemeier R, Hulsch R, Metzger JO, Berthe-Corti L (2003) Submersed culture production of extracellular wax esters by the marine bacterium Fundibacter jadensis. Mar Biotechnol 5:579–583
- Bruno L, Di Pippo F, Antonaroli S, Gismondi A, Valentini C, Albertano P (2012) Characterization of biofilm-forming cyanobacteria for biomass and lipid production. J Appl Microbiol 113:1052–1064.
- Pádrová K, Lukavsky J, Nedbalová L, Čejková A, Cajthaml T, Sigler K, Vítová M, Řezanka T (2015) Trace concentrations of iron nanoparticles cause overproduction of biomass and lipids during cultivation of cyanobacteria and microalgae. J Appl Phycol 27:1443–1451.
- Karatay SE, Dönmez G (2011) Microbial oil production from thermophile cyanobacteria for biodiesel production. Appl Energy 88:3632–3635.
- Barney BM, Wahlen BD, Garner E, Wei J, Seefeldt LC (2012) Differences in substrate specificities of five bacterial wax ester synthases. Appl Environ Microbiol 78:5734–5745.
- Lenneman EM, Ohlert JM, Palani NP, Barney BM (2013) Fatty alcohols for wax esters in Marinobacter aquaeolei VT8: two optional routes in the wax biosynthesis pathway. Appl Environ Microbiol 79:7055–7062.
- Bryn K, Jantzen E, Bøvre K (1977) Occurrence and patterns of waxes in Neisseriaceae. J Gen Microbiol 102:33–43.
- Francisco EC, Franco TT, Wagner R, Jacob-Lopes E (2014) Assessment of different carbohydrates as exogenous carbon source in cultivation of cyanobacteria. Bioprocess Biosyst Eng 37:1497–1505.
- Modiri S, Sharafi H, Alidoust L, Hajfarajollah H, Haghighi O, Azarivand A, Zamanzadeh Z, Zahiri HS, Vali H, Noghabi KA (2015) Lipid production and mixotrophic growth features of cyanobacterial strains isolated from various aquatic sites. Microbiology 161:662–673.
- Bharti RK, Srivastava S, Thakur IS (2014a) Production and characterization of biodiesel from carbon dioxide concentrating chemolithotrophic bacteria, Serratia sp. ISTD04. Bioresour Technol 153:189–197.
- Kaiwan-arporn P, Dong Hai P, Thi Thu N, Annachhatre AP (2012) Cultivation of cyanobacteria for extraction of lipids. Biomass Bioenerg 44:142–149.
- Patel VK, Maji D, Singh AK, Suseela MR, Sundaram S, Kalra A (2014) A natural plant growth promoter, calliterpenone, enhances growth and biomass, carbohydrate, and lipid production in cyanobacterium Synechocystis PCC 6803. J Appl Phycol 26:279–286.
- Okamura Y, Nakai S, Ohkawachi M, Suemitsu M, Takahashi H, Aki T, Matsumura Y, Tajima T, Nakashimada Y, Matsumoto M (2016) Isolation and characterization of bacterium producing lipid from short-chain fatty acids. Biores Technol 201:215–221.
- Bharti RK, Srivastava S, Thakur IS (2014b) Extraction of extracellular lipids from chemoautotrophic bacteria Serratia sp. ISTD04 for production of biodiesel. Bioresour Technol 165:201–204. (doi:10.1016/j.biortech.2014.02.075)
- De Andrès C, Espuny MJ, Robert M, Mercadé ME, Manresa A, Guinea J (1991) Cellular lipid accumulation by Pseudomonas aeruginosa 44T1. Appl Microbiol Biotechnol 35:813–816.
- Patnayak S, Sree A (2005) Screening of bacterial associates of marine sponges for single cell oil and PUFA. Lett Appl Microbiol 40:358–363.
- Alvarez HM, Steinbüchel A (2002) Triacylglycerols in prokaryotic microorganisms. Appl Microbiol Biotechnol 60:367–376.
- Lennen RM, Pfleger BF (2012) Engineering Escherichia coli to synthesize free fatty acids. Trends Biotechnol 30:659–667.
- Lu X, Vora H, Khosla C (2008) Overproduction of free fatty acids in E. coli: implications for biodiesel production. Metab Eng 10:333–339.
- Liu T, Vora H, Khosla C (2010) Quantitative analysis and engineering of fatty acid biosynthesis in E. coli. Metab Engin 12:378–386.
- Zhang X, Li M, Agrawal A, San KY (2011) Efficient free fatty acid production in Escherichia coli using plant acyl-ACP thioesterases. Metab Eng 13:713–722.
- Ranganathan S, Wei Tee T, Chowdhury A, Zomorrodi AR, Moon Yoon J, Fu Y, Shanks JV, Maranas CD (2012) An integrated computational and experimental study for overproducing fatty acids in Escherichia coli. Metab Engin 14:687–704.
- Liu H, Yu C, Feng D, Cheng T, Meng X, Liu W, Zou H, Xian M (2012) Production of extracellular fatty acid using engineered Escherichia coli. Microb Cell Fact 11:41–54.
- Zheng Y, Li L, Liu Q, Qin W, Yang J, Cao Y, Jiang X, Zhao G, Xian M (2012) Boosting the free fatty acid synthesis of Escherichia coli by expression of a cytosolic Acinetobacter baylyi thioesterase. Biotechnol Biofuels 5:76–88.
- Fan L, Liu J, Nie K, Liu L, Wang F, Tan T, Deng L (2013) Synthesis of medium chain length fatty acid ethyl esters in engineered Escherichia coli using endogenously produced medium chain fatty acids. Enzyme Microb Tech 53:128–133.
- Wu H, Karanjikar M, San KY (2014a) Metabolic engineering of Escherichia coli for efficient free fatty acid production from glycerol. Metab Engin 25:82–91.
- Wu H, Lee J, Karanjikar M, San KY (2014b) Efficient free fatty acid production from woody biomass hydrolysate using metabolically engineered Escherichia coli. Biores Technol 169:119–125.
- Cao Y, Liu W, Xu X, Zhang H, Wang J, Xian M (2014) Production of free monounsaturated fatty acids by metabolically engineered Escherichia coli. Biotechnol Biofuels 7:59–70.
- Kurosawa K, Wewetzer SJ, Sinskey AJ (2014) Triacylglycerol production from corn stover using a xylose-fermenting Rhodococcus opacus strain for lignocellulosic biofuels. J Microb Biochem Technol 6:254–259.
- Röttig A, Zurek PJ, Steinbüchel A (2015) Assessment of bacterial acyltransferases for an efficient lipid production in metabolically engineered strains of E. coli. Metab Engin 32:195–206.
- Hernández MA, Comba S, Arabolaza A, Gramajo H, Alvarez HM (2015) Overexpression of a phosphatidic acid phosphatase type 2 leads to an increase in triacylglycerol production in oleaginous Rhodococcus strains. Appl Microbiol Biotechnol 99:2191–2207.
- Kurosawa K, Wewetzer SJ, Sinskey AJ (2014) Engineering xylose metabolism in triacylglycerol producing Rhodococcus opacus for lignocellulosic fuel production. Biotechnol Biofuels 6:134–147.
- Kurosawa K, Laser J, Sinskey AJ (2015c) Tolerance and adaptive evolution of triacylglycerol-producing Rhodococcus opacus to lignocellulose-derived inhibitors. Biotechnol Biofuels 8:76–90.
- Kurosawa K, Plassmeier J, Kalinowski J, Rückert C, Sinskey AJ (2015a) Engineering L-arabinose metabolism in triacylglycerol-producing Rhodococcus opacus for lignocellulosic fuel production. Metab Engin 30:89–95.
- Kurosawa K, Radek A, Plassmeier JK, Sinskey AJ (2015b) Improved glycerol utilization by a triacylglycerol producing Rhodococcus opacus strain for renewable fuels. Biotechnol Biofuels 8:31–42.
- Rucker J, Paul J, Pfeifer BA, Lee K (2013) Engineering E. coli for triglyceride accumulation through native and heterologous metabolic reactions. Appl Microbiol Biotechnol 97:2753–2759.
- Lin F, Chen Y, Levine R, Lee K, Yuan Y, Lin XN (2013) Improving fatty acid availability for bio-hydrocarbon production in Escherichia coli by metabolic engineering. PLoS ONE 8:e78595.
- Janßen HJ, Steinbüchel A (2014) Production of triacylglycerols in Escherichia coli by deletion of the diacylglycerol kinase gene and heterologous overexpression of atfA from Acinetobacter baylyi ADP1. Appl Microbiol Biotechnol 98:1913–1924.
- Comba S, Sabatini M, Menendez-Bravo S, Arabolaza A, Gramajo H (2014) Engineering a Streptomyces coelicolor biosynthesis pathway into Escherichia coli for high yield triglyceride production. Biotechnol Biofuels 7:172–183.
- Kalscheuer R, Stölting T, Steinbüchel A (2006) Microdiesel: Escherichia coli engineered for fuel production. Microbiology (SGM) 152:2529–2536.
- Elbahloul Y, Steinbüchel A (2010) Pilot-scale production of fatty acid ethyl esters by an engineered Escherichia coli strain harboring the p(Microdiesel) plasmid. Appl Environ Microbiol76:4560–4565.
- Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, del Cardayre SB, Keasling JD (2010) Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463:559–562.
- Duan Y, Zhi Z, Ke C, Xiaoming T, Xuefeng L (2011) De novo biosynthesis of biodiesel by Escherichia coli in optimized fed-batch cultivation. PLoS ONE 6:1–7.
- Zhang F, Carothers JM, Keasling JD (2012) Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat Biotechnol 30:354–359.
- Bokinsky G, Peralta-Yahya PP, George A, Holmes BM, Steen EJ, Dietrich J, Lee TS, Tullman-Ercek D, Voigt CA, Simmons BA, Keasling JD (2011) Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli. PNAS 108:19949–19954.
- Guo D, Zhu J, Deng Z, Liu T (2014) Metabolic engineering of Escherichia coli for production of fatty acid short-chain esters through combination of the fatty acid and 2-keto acid pathways. Metab Engin 22:69–75.
- Tao H, Guo D, Zhang Y, Deng Z, Liu T (2015) Metabolic engineering of microbes for branched-chain biodiesel production with low-temperature property. Biotechnol Biofuels 8:92–103.
Related Resources
Lipid Library
Edible Oil Processing
In the present context, the term edible oil processing covers the range of industrial…
Lipid Library
The Highs and Lows of Cannabis Testing
October 2016 With increasing legalization of both adult recreational and medical cannabis,…
Lipid Library
The secrets of Belgian chocolate
By Laura Cassiday May 2012 Like a bonbon nestled snugly in a…