Weaving together genetics, epigenetics, and the microbiome to optimize human nutrition
By Rebecca Guenard
October 2019
- The human body holds the answers to its own optimal dietary needs, but can scientists decipher all the clues encompassing personalized nutrition to maximize our chances of a long life with the best possible health?
- Studies on identical twins diminish genetics’ dominance in human health and highlight the importance of lifestyle choices.
- A better understanding of human populations and the function of the microbiome may help clarify the interaction between genetics and environment in human nutrition.
- Emerging evidence supports the notion that nutrition will be personalized in the future, since no one diet suits everybody.
Like most of us, when Bryan eats potato chips his blood triglyceride levels spike, but when his twin brother eats the same chips his triglyceride levels are six times those of his genetic equivalent. When the Human Genome Project was launched in the early 1990s, scientists assumed that identifying our genes would reveal places in our DNA that can be manipulated for peak health. However, after four decades of research, the body’s response to food has proven to be more complicated than they thought.
“We thought that genetics would explain everything,” says Iwona Rudkowska, an associate professor at Laval University in Québec, Canada. “People thought, ‘If we find out our genetics, then we will know exactly what we should eat. This is not the case.”
Health is influenced by more than just our genes. Environmental factors, such as sleep and stress, can influence how genes function and affect our body’s response to food. Now, more and more experts are stating that to improve public health, nutritional science needs to widen its scope to include details about individual responses to food.
Current dietary recommendations are based on the outcome of studies involving thousands of participants. “The outcome is the mean; how the average person responds,” says Rudkowska. “But when you look at the data, some individuals respond completely differently than others.”
The field of nutrition is now aiming for personalization. The goal is to deliver healthful eating guidelines tailored to the genetic, epigenetic, metabolomic, and microbiomic specifications of each individual. Large, long-term studies, like PREDICT and Food4Me are filling massive databases so computers can establish models that will forecast the best diet for an individual based on that person’s unique parameters. Rudkowska says, “People are starting to realize that we are not all the same.”
Stratification before personalization
Personalized nutrition stems from the notion of personalized medicine. The efficacy of certain cancer treatments, for example, are known to be influenced by a person’s DNA. However, just as personalized medicine is not as turnkey as was once hoped, nutrition researchers are finding that genetic factors explain only a fraction of an individual’s response to food.
For the past 10 years, Rudkowska has performed genome-wide association studies (GWAS) to evaluate if the effectiveness of omega-3 supplements are dependent on a genetic component. Her experiments typically involve hundreds of participants of Canadian decent whose plasma triglyceride levels are measured over several weeks while taking a supplement containing eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA).
Rudkowska initially found four genes (IQCJ, NXPH1, PHF17, and MYB) that seemed to influence the body’s response to omega-3s (https://doi.org/10.3390/ijms18020257). In other experiments, her group then identified 13 locations where the base pair of an individual whose triglycerides were lowered by omega-3s was different from individuals with no response (https://doi.org/10.1194/jlr.M045898). And, with finer genome mapping, her most recent experiments reveal 31 locations on the genome associated with the lowering of triglycerides (https://doi.org/10.1093/ajcn/nqy298).
Although her group has homed in on these potential genetic risk factors, Rudkowska says they cannot be used as personalized nutrition targets. “Even if we have a recommendation that a certain food will decrease cholesterol or LDL, that may not be true for everybody,” she says. “The science is not there yet.”
When glucose, insulin, or triglyceride blood levels are measured to study a person’s response to a food or treatment, researchers find a wide variety of results. Some participants have a rapid and prolonged increase in blood sugar and insulin; others experience a spike in fat levels that lingers long after a meal. Either response could signal future problems like weight gain and diabetes or heart disease. However, when attempting to connect these blood results to a person’s DNA, scientists have not been able to produce a strong correlation. Genetic factors account for less than 50% of a person’s response to glucose. For insulin, it is less than 30%, and for triglycerides its under 20% (https://tinyurl.com/yyw3gbu8).
“We need to do more research before we can make exact recommendations on foods that one person would respond to better than another,” says Rudkowska. For now, the best researchers can do with genetic data is to stratify people. Researchers who work in stratified nutrition consider dietary recommendations for specific groups, such as children, diabetics, or pregnant women, instead of individuals. “We know that individuals who have a higher risk of cardiovascular disease probably have higher triglycerides, so they might react better to omega-3s,” she says. “We categorize it that way, but we are not at the point yet where we could use the genetic information, because there are so many other uncertainties.”
Rudkowska is hoping that modeling will reduce some of the uncertainty and better predict responses. In parallel with her triglyceride experiments, she is studying the diverse effects that have been found between dairy consumption and blood glucose levels. Some researchers have identified effects of consuming dairy for individuals with type 2 diabetes, while other have reported that dairy is detrimental to those with the disease (https://doi.org/10.1093/cdn/nzz083). Her group is collaborating with researchers who do bioinformatics to determine if they can find a biological source for the variability.
“We gathered data on microbiome, epigenetic, and metabolic factors. Then we used machine learning to develop an algorithm for predicting a person’s response,” Rudkowska says. With just one such factor, they can predict how dairy intake will affect the blood glucose level of a study participant with an accuracy of 65–70%. She says they hope to construct a better algorithm by incorporating multiple factors.
“I think we are moving away from just studying one gene and one food because it is too simplistic,” says Rudkowska. “It is nice to think that we will react a certain way because of a genetic variation, but that is not what is happening. One person who has this variation might respond completely differently from someone else with the same variation.” The results of ongoing studies with twins reiterate this fact. Nutritional studies now involve extensive data sets comparing every aspect of the lives of two people with the same genetics, yet different health outcomes.
Twins and epigenetics
In 1992, Tim Spector, professor of genetic epidemiology at King’s College in London, UK, started analyzing biological parameters of identical twins to see if he could uncover a genetic reason for osteoporosis and other rheumatologic diseases (www.twinsuk.ac.uk). He started out with a cohort of a few hundred participants that bloomed into a registry of 12,000 twins in the UK. Spector and his team collected blood, urine, and tissue from the twins, who were mostly middle-aged women, with equal numbers of identical and fraternal twins.
Over the decades, the program has provided a trove of genetic information on healthful aging and complex diseases. The data stored in the TwinsUK database is available to the scientific community, and it has prompted extensive research on everything from cardiovascular and metabolic disorders to behavioral and socio-economic characteristics of the population. This is due to the database being a motherload of systems biology resources: genome-wide scans of single nucleotide variants, RNA sequencing, metabolic profiles, gut flora microbiomics, epigenetic markers, and gene expression arrays, to name a few.
Some of the most intriguing results to arise from TwinsUK, reveal how lifestyle has a direct effect on gene expression. Genes are regulated by molecules that piggyback onto DNA and determine if the gene participates in the functions of the human body. This system is known as epigenetics.
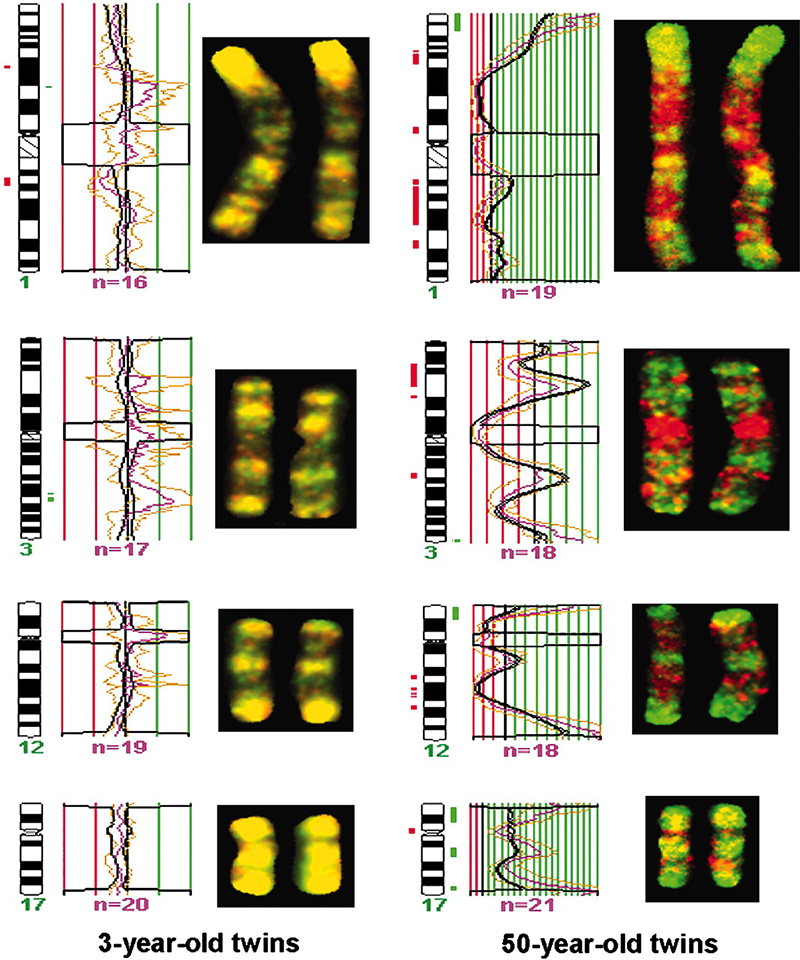
Spector and his team reported in a 2005 paper that identical twins are epigenetically indistinguishable during their early years of life. However, when they compared the genetic information of 3-year-old identical twins to 50-year-old identical twins, they found that the epigenetics of the older twins were notably different (https://doi.org/10.1073/pnas.0500398102). Although each set of twins were born with the exact same DNA, the expression of their genes diverged as they aged and experienced different things. Since these early, intriguing studies Spector’s team has pursued an ambitious goal of explaining the human’s vast variability in response to diet.
FIG. 1. Chromosomal regions of 3- and 50-year-old identical twins showing the epigenetic change of DNA methylation. Green and red areas show changes on the chromosome where methylation occurred, whereas the 3-year-old twins have similar methylation distribution indicated by their consistent yellow color. Source: Fraga et al., PNAS, 102, 30, 2005.
In June of 2019, the PREDICT 1 study published its initial findings and launched the second phase of the experiment, PREDICT 2 (https://predict.study/). Self-reported nutritional studies contain the inherent flaw that humans are notoriously bad at remembering what they ate. The PREDICT study avoided this by giving each participate a full clinical work-up prior to starting. Everyone then ate standardized meals in the clinic on the first day. Their blood was drawn 10 times and measured for sugar, insulin, fat, and inflammation biomarkers, among others. For the following two weeks, participants ate their own meals along with standardized foods. A wearable monitor recorded their blood sugar level every 15 minutes, and they took regular blood samples. Data collection on the participants was quite comprehensive, including monitored sleep and activity patterns, as well as regular stool samples.
The study concluded that the genetic contribution to the body’s metabolic response is minor. There is little relevance in amount of fat or carbohydrates in a participant’s food either. According to the PREDICT 1 report, the most important determinants of a healthy metabolic response were sleep, stress, exercise, and the gut microbiome.
Gut check
As scientists dig through the ecosystem of the human intestinal tract attempting to parse out the role of billions of microbes, reports on new insights seem never-ending. Most recently, a team at Stanford University School of Medicine in Palo Alto, California, USA, discovered that the microbiome produces thousands of small proteins that were previously unknown and whose purpose remains to be understood (https://tinyurl.com/yyzcb4pz). As scientists unearth a constant flux of information about the microscopic landscape within the gut, each new find lends credence to the growing acknowledgement that nutritional choices affect the bacterial population ensconced there.
“The microbiota have many established roles in promoting host health,” says Lauren Rajakovich, postdoc in the laboratory of Emily Balskus at Harvard University in Boston, Massachusetts, USA. She says there is still many unanswered questions, but mouse models designed with no microbiome relay the importance of these intestinal colonies.
“These model systems often have an underdeveloped immune system, so they are susceptible to different pathogens,” she says. “They need a lot of supplementation in their diets.” Without a microbiome, such mice cannot maintain good health. Scientists have established that the microbiome is needed for the biosynthesis of specific molecules, like vitamins, that are not formed anywhere else in the host’s digestive system. Through these mouse models, researchers have determined that gut bacteria provide a last-ditch source of energy and nutrients as food waste exits the body.
“Food components that the human body is not able to digest end up in our large intestine, and the microbes that live there are able to breakdown these complex polysaccharides into smaller units that are reabsorbed by the host,” Rajakovich says. She adds that the carbohydrates are reduced to simple sugars that the microbes convert into short-chain fatty acids used as an energy source by epithelial cells in the large intestine.
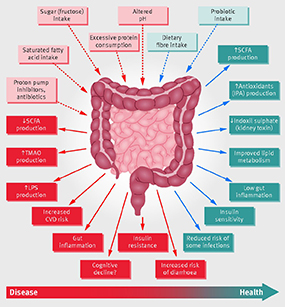
FIG. 2. Schematic representation of the role of the gut microbiota in health and disease giving some examples of inputs and outputs. CVD=cardiovascular disease; IPA=indolepropionic acid; LPS=lipopolysaccharide; SCFA=short chain fatty acid; TMAO=trimethylamine N-oxide.Source: Valdes et al., BMJ, 361, k2179, 2018.
The Balskus lab deciphers common metabolic pathways within the microbiome by concentrating on small molecule chemistry. Gut bacteria metabolize the contents of the intestines for their own nutritional and energy needs. Sometimes the small molecules they produce benefit the host by, for example, producing molecules that regulate immune responses, but sometimes gut bacteria dispel molecules that cause disease.
“Our lab is really excited about exploring the production of trimethylamine by the gut microbiome, because it has not only been associated primarily with cardiovascular disease, but also with chronic kidney disease, nonalcoholic fatty liver disease, and systemic things, like obesity and diabetes,” Rajakovich says. The small molecule trimethylamine (TMA) is not made anywhere in the body except in the gut. Intestinal microbes convert choline from a variety of foods, such as, red meat, eggs, soy, or dairy, into TMA that is converted to trimethylamine oxide (TMAO) in the liver. TMAO is linked to plaque buildup and cardiovascular disease. The Balskus research group is exploring whether in the future a therapeutic could be administered to patients to reroute this metabolic pathway away from disease.
Many microbiome studies focus on bacterial metabolic pathways, with an eye toward treatment. For example, prebiotics have become a popular way to ramp up beneficial bacteria. A fiber-rich diet is known to promote an optimal ratio for glucose-regulating bacteria.
Details of various aspects of the microbiome are being discovered but designing personalized nutrition will require largescale studies that incorporate environmental as well as dietary factors. “We often focus in on a particular bacterium and sometimes we do forget that they are part of this much larger and complex community that is all interconnected,” says Rajakovich. She adds that the study of human gut microbiota would really benefit if ecologists and population geneticists worked together to figure out such complex systems, while chemists and biochemists continue to explore the metabolism.
The pocket nutritionist
The implementation of high-throughput tools for genomics, microbiomics, and metabolomics, together with the computational capabilities of artificial intelligence (AI), have led to the opportunity to create a customized nutritional plan for anyone. Just as 23andMe and AncestryDNA offer to give customers a glimpse into their heritage, a similar industry is forming around personalized nutrition.
The Santa Clara, California-based microbiome analysis company, Viome, launched in 2019 with the ambition to “take the guesswork out of eating right with a science-based, at-home microbiome test,” (https://www.viome.com/). Viome analyzes the genes expressed by the microbiome, labels them good or bad, and advises customers about foods that will improve their health. Spector has launched a product based on his TwinsUK research. His app for your phone, ZOE, describes the best diet for a user’s metabolism (https://joinzoe.com/).
The interesting thing about both products is that customer information will contribute to a database that will use AI to better predict personalized nutrition for future users. The more people purchase the products, the more data gets added to the AI algorithm and, presumably, the better it will be at guessing a consumer’s specific dietary needs. Such big data establishes a predictive model that can be used to evaluate an individual’s response to a nutritional intervention, while also providing nutrition experts with an understanding of how diet influences health and disease.
“One caveat with this type of product is that what we understand about the gut microbiome is that it is very flexible and temporal,” Rajakovich. “The difference between that and 23andMe is that your genetics is never changing.” Rudkowska says researchers still have a lot to resolve concerning the microbiome, but these big data studies are the future of nutrition. She says the technology to analyze all the body’s biological parameters is now available; the data is easy to acquire. The challenge is how to interpret all the information researchers collect.
“These studies are complicated,” she says. “But the human body is complicated.”
References
- Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity, Chambers, et al., Nature 541: 7635, 2017.
- Genetic determinants of the gut microbiome in UK twins, Goodrich, et al., Cell Host & Microbe 19: 731, 2016.
- Heritable components of the human fecal microbiome are associated with visceral fat, Beaumont, et al., Genome Biol. 17: 189, 2016.
- Systems biology approaches to understand the effects of nutrition and promote health, Badimon, et al., Br. J. Clin. Pharmacol. 83: 38, 2017.
- Personalized nutrition and health, Orvodas, et al., BMJ 361: k2173, 2018.
- Paving the way to precision nutrition through metabolomics, Tebani and Bekri, Frontiers in Nutrition 6: 41, 2019.
- Addressing the nutritional phenotype through personalized nutrition for chronic disease prevention and management, Laddu and Hauser, Progress in Cardiovascular Diseases 62: 9, 2019.
- Metabolic functions of the human gut microbiota: the role of metalloenzymes, Rajakovich and Balskus, Nat. Prod. Rep. 36: 593, 2019.
Feeding the skin microbiome Dermatologists have understood for a long time that bacteria on the skin’s surface irritate hair follicles that cause acne, which is routinely combatted with prescription antibiotics. Just as antibiotics in the human gut eliminate good bacteria, most acne treatments do the same thing in skin.
The latest research suggests that instead of annihilating bacterial colonies, skin treatments should foster an environment for growth. Beauty companies are now taking stock of the skin’s microorganisms to evaluate how best to promote skin health and treat a range of conditions.
Billions of microorganisms, including bacteria, fungi, mites, and viruses, can stake claim to a single square centimeter of skin, but bacteria are considered the most useful. They are present on the surface and in deeper layers, indirectly protecting the skin against pathogens by competing for resources, and directly by producing antimicrobial compounds—some of which release anti-inflammatory compounds. Not all bacteria share the same list of attributes, however, so cosmetic companies are trying to differentiate and promote the superstars.
As is the case with the gut, the science of the skin’s microbiome is still being established, but beauty companies are already using the concept in new product lines, especially for the treatment of acne. Earlier this year, Stat News reported on two start-ups that are betting on the benefits of the microbiome (https://tinyurl.com/y4l24tfe). Both companies were launched within a Johnson & Johnson tech incubator located in San Francisco, California. One of the start-ups, Naked Biome, sequenced a strand of bacteria that is present on people with healthy skin and developed a cleansing pad soaked in the specific microbe. They are currently gathering data on the product for a future FDA filing. The other start-up, Ellis Day, is creating a serum containing three bacteria-killing viruses that they plan to market as an over-the-counter product.
With time, beauty companies will know if consumer success can be gleaned from a skincare product made from live viruses and bacteria. But companies like Johnson and &Johnson and Henkel have identified the microbes populating our skin as beauty industry game-changers and will continue to put scientific resources into determining how to use them to their advantage.