Omics reveals subtle changes in carbon flux that lead to increased oil biosynthesis in oil palm
By David Ross Appleton, Teh Huey Fang, Neoh Bee Keat, Ooi Eng Keong, Wong Yick Ching, and Harikrishna Kulaveerasingam
May 2014
- Detailed biochemical analysis during plant development can help bridge the gaps between biological understanding, genetic markers, and phenotypes.
- Despite significant experimental design and interpretation challenges in a field environment for a crop such as oil palm, useful insights can be gained by conducting biochemical studies.
- Concerted changes in nitrogen metabolites to support oil biosynthesis and fruit development along with flux differences in glycolysis were observed to contribute to large increases in overall yield.
Oil palm is the most productive oil-producing crop. On a commercial basis, yields of 4–6 metric tons of oil per hectare (MT oil/ha) are produced, while yields of up to 12 MT oil/ha have been achieved in breeding trials. With yields of up to 17 MT oil/ha theoretically possible, there is much potential for oil palm to meet increased demands for biofuels and edible oils into the future without increasing new plantings.
A large proportion of the oil palm’s high productivity is due to its being a perennial and the tropical environment in which it thrives. However, the oil biosynthetic machinery that produces up to 90% oil in the fruit mesocarp is also unique.
The development of genetic markers for improved selection and breeding has been reported increasingly in recent years, culminating in the publication of the oil palm genome in 2013 by a consortium including researchers with the Malaysian Palm Oil Board, Orion Genomics (St. Louis, Missouri, USA), Washington University (St. Louis, Missouri), Cold Spring Harbor Laboratory (Cold Spring Harbor, New York, USA), the University of Arizona (Tucson, USA), and the Sackler Institute for Comparative Genomics (New York City, New York, USA). As achieved in other crops, the use of genetic markers for oil palm crop improvement holds much promise. However, marker accuracy varies across population backgrounds depending on genetic distance of markers from controlling genes and population diversity. In addition, the development of accurate marker selection tools is especially difficult for traits that are highly polygenic and have large environment effects, such as yield .
Biochemical research using omics and genetic marker approaches
Biochemical omics techniques can serve as a bridge between controlling genes and associated genetic markers by identifying genes and biochemical pathways that are more directly associated with the final trait. At Sime Darby (a Malaysia-based multinational conglomerate involved in, among other things, oil palm plantations and palm oil production), we took the approach of investigating the differences in biochemical regulation that distinguish highest-yielding individuals (10–12 MT oil/ha) from average performers (4–7 MT oil/ha) within a group of siblings planted in the same location. Biochemical changes before, during, and after oil biosynthesis in fruit mesocarp were compared using transcriptomics, proteomics, and metabolomics and provided insight into oil biosynthesis control in the oil palm. Genome polymorphisms surrounding identified genes were then prioritized when conducting large-scale genome-wide association studies on larger sets of more diverse populations.
Challenges in conducting omics research in a field crop
As a perennial tree crop, the oil palm poses significant challenges for experimental design and interpretation when attempting to relate the short-term microscale cellular processes to macroscale long-term yield. While the logistics of sampling and preservation of biochemicals that change owing to enzymatic action soon after sampling is a key issue, this can be addressed through immediate washing and freezing of samples using liquid nitrogen in the field.
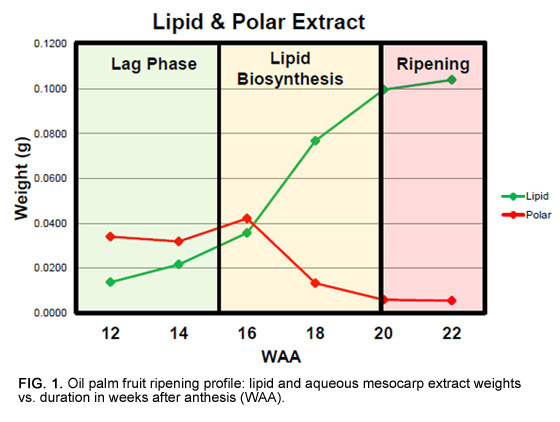
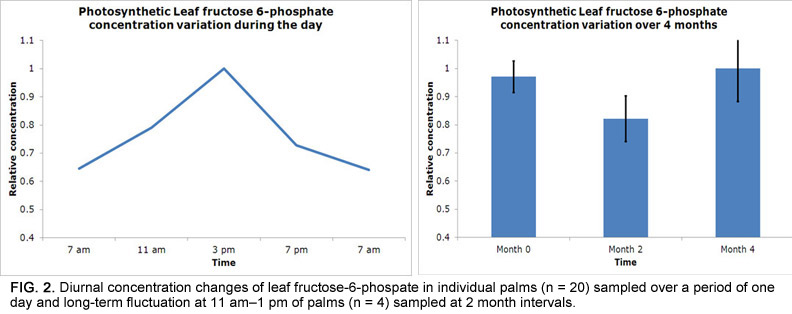
A more significant issue is the extremely long duration over which samples must be collected due to the destructive nature of fruit sampling. Since one bunch needs to be sampled for each time point, sampling of the 16 palms in this study took two years to complete and required six bunches per palm to be tracked from pollination until sampling between 12 and 22 weeks after anthesis (WAA; Fig. 1). Given that biochemical levels and gene expression vary greatly owing to environmental conditions, this was a significant concern. For example, there are significant changes in leaf metabolite levels over time, whereby sugar phosphates (precursors for fatty acid biosynthesis) vary more throughout the day than over a period of months in the same palm (Fig. 2). Fruit metabolite levels are expected to be more stable, and we confirmed that observed temporal trends were in line with expectations before further result interpretation.
Omics results reflect drivers of yield
When studied between 12 and 22 WAA, protein concentrations and gene expression levels were consistent with metabolite changes in many cases. The three datasets were simplified by categorizing the genes, proteins, and metabolites into their cellular pathways and identifying areas of concordance. Several global trends were evident in the metabolomic results; for example, high-yielding palms have significantly higher levels of amino acids before oil biosynthesis commences, but the levels decrease to the same as average-yielding palms once the fruits are ripe.
The higher accumulation of these nitrogen-containing protein building blocks is possibly related to fueling future enzyme production and mesocarp growth. Nucleosides were generally observed to increase along with lipid concentration, with high-yielding palms again showing significantly higher levels. These data highlight the important role of nitrogen assimilation for oil production and indicate multiple processes are required to support oil production aside from lipid biosynthesis alone.
The role and regulation of these biosynthetic pathways will be a key focus in future studies. Metabolites that are more directly linked to oil biosynthesis in glycolysis and the tricarboxylic acid (TCA) cycle also exhibited differentials in high-yielding palms. We observed an apparent change in flux at a specific branch-point in the middle of the glycolytic pathway, involving four key enzymes: fructose-1,6-biphosphate aldolase, triosephosphate isomerase, glycerol-3-phosphate dehydrogenase, and glyceraldehyde-3-phosphate dehydrogenase. Concordance with protein and RNA levels was evident and provided confidence in the significance of this group of enzymes. Differentials indicated that high-yielding palms divert more carbon toward the biosynthesis of glycerol than lower yielding palms (Fig. 3).
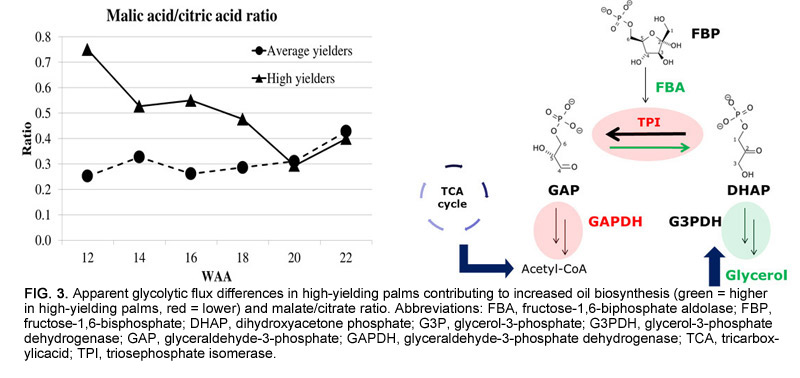
The ability to increase the levels of glycerol in cells has been previously linked in other oil crops to significantly higher yields due to its role as a lipid building block. Significant differentials were also observed in certain metabolites in the TCA cycle. The ratio of malate to citrate changed markedly during lipid biosynthesis, and high-yielding palms had a much higher ratio throughout. The energy carrier molecule ATP had lower levels in high-yielding palms during oil biosynthesis, possibly reflecting the increased demand of energy for fatty acid production, while several proteins involved in ATP metabolism were notably up-regulated using iTRAQ (isobaric tag for relative and absolute quantitation) protein analysis.
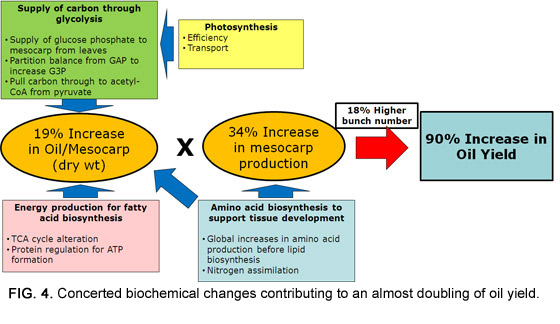
Taken together, the omics analysis of oil palm mesocarp enabled the identification of genes that play important roles leading to increased oil production. The patterns observed were in agreement with phenotypic data obtained through detailed yield records conducted over seven years prior to the biochemical analysis. Data for yields revealed simultaneous increases in the number of bunches produced per year, the average mesocarp mass per bunch, and the oil content of the mesocarp; while omics analysis indicated changes in glycolytic flux, nitrogen assimilation, and energy management (Fig. 4).
Subsequently, we have confirmed in yeast systems the effect on oil biosynthesis of several of these genes and identified over 60,000 DNA variants (SNPs; also known as single nucleotide polymorphisms) in the sequences of the ~2,000 differential genes identified. The identified SNPs were prioritized out of more than three million detected throughout the oil palm genome for the purpose of genome-wide association studies, since it was not economically feasible to genotype using all. This led to an analysis based on 200,000 SNPs across the genome with increased density near differential genes. This has enabled the identification of alleles of specific genes that can be associated with traits across the wider populations of oil palm in our collection.
The authors thank Chew Fook Tim (National University of Singapore) for consultation and their colleagues at Sime Darby Technology Centre and Sime Darby Research for their significant contributions to sample/data acquisition and discussions.
David Ross Appleton, Teh Huey Fang, Neoh Bee Keat, Ooi Eng Keong, Wong Yick Ching, and Harikrishna Kulaveerasingam are with Sime Darby. They can be contacted at their respective email addresses: david.ross.appleton@simedarby.com, teh.huey.fang@simedarby.com, neoh.bee.keat@simedarby.com, tony.ooi.engkeong@simedarby.com, wong.yick.ching@simedarby.com, and harikrishna.k@simedarby.com.
Information
Bourgis, F., A. Kilaru, X. Cao, G.F. Ngando-Ebongue, N. Drira, J.B. Ohlrogge, and V. Arondel, Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning, Proc. Natl. Acad. Sci. USA 108:12527–12532 ,2011.
Dussert, S., C. Guerin, M. Andersson, T. Joet, T.J. Tranbarger, M. Pizot, G. Sarah, A. Omore, T. Durand-Gasselin, and F. Morcillo, Comparative transcriptome analysis of three oil palm fruit and seed tissues that differ in oil content and fatty acid composition, Plant Physiol. 162:1337–1358, 2013.
Loei, H., J. Lim, M. Tan, T.K. Lim, Q.S. Lin, F.T. Chew, H. Kulaveerasingam, and M.C.M. Chung, Proteomic analysis of the oil palm fruit mesocarp reveals elevated oxidative phosphorylation activity is critical for increased storage oil production, J. Proteome Res. 12:5096–5109, 2013.
Neoh, B.K., H.F. Teh, T.L.M. Ng, S.H. Tiong, Y.M. Thang, M.A. Ersad, M. Mohamed, F.T. Chew, H. Kulaveerasingam, and D.R. Appleton, Profiling of metabolites in oil palm mesocarp at different stages of oil biosynthesis, J. Agric. Food Chem. 61:1920–1927, 2013.
Rajinder, S., M. Ong-Abdullah, E.-T.L. Low, M.A.A. Manaf, R. Rosli, et al., 2013, Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds, Nature 500:335–339, 2013.
Teh, H.F., B.K. Neoh, M.P.L. Hong, J.Y.S. Low, T.L.M. Ng, N. Ithnin, Y.M. Thang, M. Mohamed, F.T. Chew, H.M. Yusof, H. Kulaveerasingam, and D.R. Appleton, Differential metabolite profiles during fruit development in high-yielding oil palm mesocarp, PLoS ONE 8(4):e61344 (2013).
